Probe CUST_2833_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2833_PI426222305 | JHI_St_60k_v1 | DMT400043948 | ATAAGTAGGAGGCAATCAAGGCAAATGTTTGAAATTGAATCAGGGATTGATCTTGAACAG |
All Microarray Probes Designed to Gene DMG400017055
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3323_PI426222305 | JHI_St_60k_v1 | DMT400043945 | CCATAATATGTTGGGTTTTCTAGCTTTTTTCTAACAGCTGATCTAGAAGATCATATGCCT |
| CUST_2833_PI426222305 | JHI_St_60k_v1 | DMT400043948 | ATAAGTAGGAGGCAATCAAGGCAAATGTTTGAAATTGAATCAGGGATTGATCTTGAACAG |
| CUST_2841_PI426222305 | JHI_St_60k_v1 | DMT400043946 | ATAAGTAGGAGGCAATCAAGGCAAATGTTTGAAATTGAATCAGGGATTGATCTTGAACAG |
Microarray Signals from CUST_2833_PI426222305
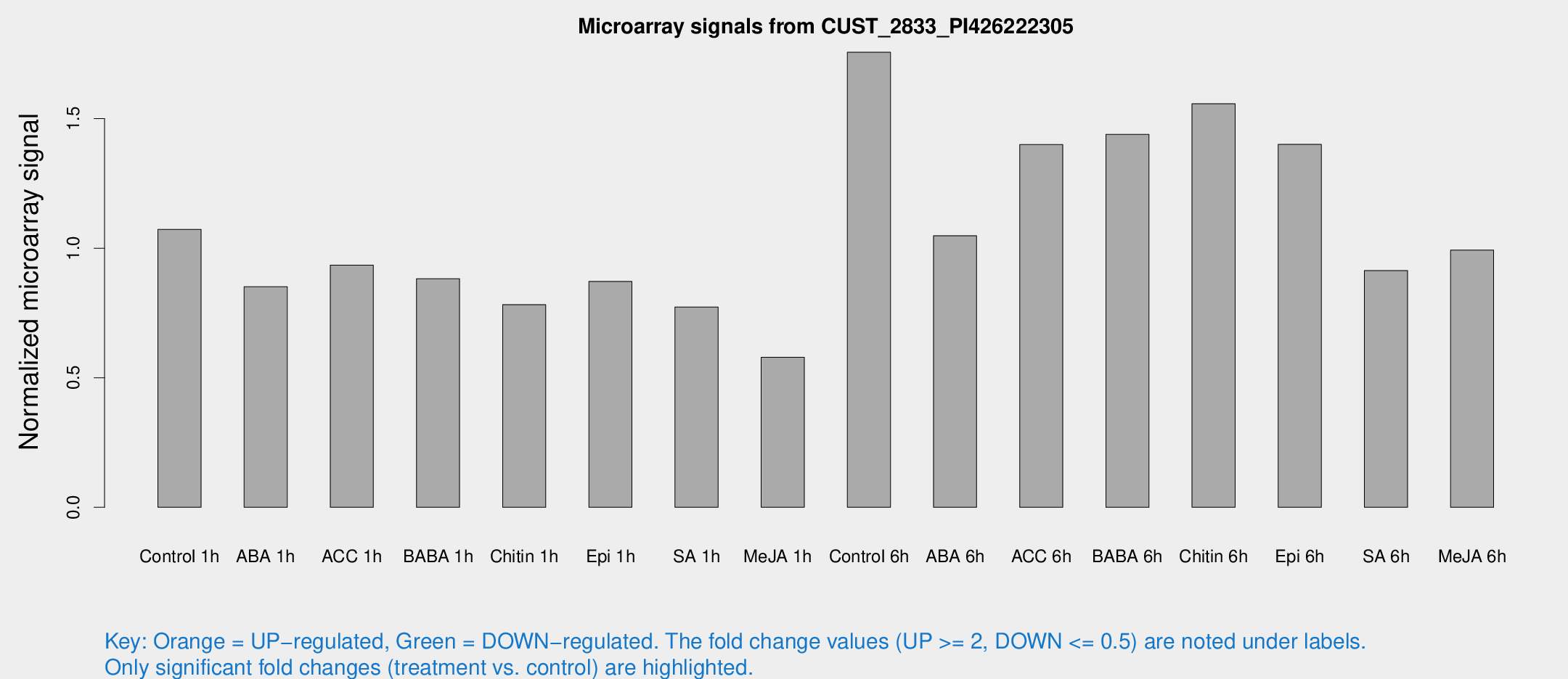
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 464.357 | 64.7445 | 1.07259 | 0.0824601 |
| ABA 1h | 323.731 | 35.6854 | 0.851072 | 0.0647439 |
| ACC 1h | 414.306 | 51.312 | 0.934685 | 0.0550356 |
| BABA 1h | 377.818 | 73.4996 | 0.882399 | 0.10406 |
| Chitin 1h | 302.811 | 34.8932 | 0.782114 | 0.0485666 |
| Epi 1h | 321.733 | 22.3335 | 0.871839 | 0.0513189 |
| SA 1h | 347.986 | 63.7792 | 0.773095 | 0.0914055 |
| Me-JA 1h | 203.542 | 25.4979 | 0.578879 | 0.0351503 |
| Control 6h | 828.09 | 251.731 | 1.75633 | 0.470193 |
| ABA 6h | 483.446 | 69.5813 | 1.04825 | 0.102618 |
| ACC 6h | 719.522 | 158.386 | 1.40005 | 0.202467 |
| BABA 6h | 718.516 | 151.177 | 1.43927 | 0.293856 |
| Chitin 6h | 714.466 | 87.5958 | 1.55722 | 0.182144 |
| Epi 6h | 681.427 | 90.0658 | 1.40063 | 0.227046 |
| SA 6h | 419.15 | 109.345 | 0.913338 | 0.18768 |
| Me-JA 6h | 470.789 | 141.593 | 0.992827 | 0.289811 |
Source Transcript PGSC0003DMT400043948 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G47560.1 | +2 | 3e-20 | 91 | 52/146 (36%) | RING/U-box superfamily protein | chr2:19511934-19512617 REVERSE LENGTH=227 |
