Probe CUST_28024_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28024_PI426222305 | JHI_St_60k_v1 | DMT400081977 | CTTCTCTTTGTACATAAGCAAGAGATATTTGTGCAATGTTCCTACTCTTCAGAATCAAAC |
All Microarray Probes Designed to Gene DMG400032183
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28024_PI426222305 | JHI_St_60k_v1 | DMT400081977 | CTTCTCTTTGTACATAAGCAAGAGATATTTGTGCAATGTTCCTACTCTTCAGAATCAAAC |
Microarray Signals from CUST_28024_PI426222305
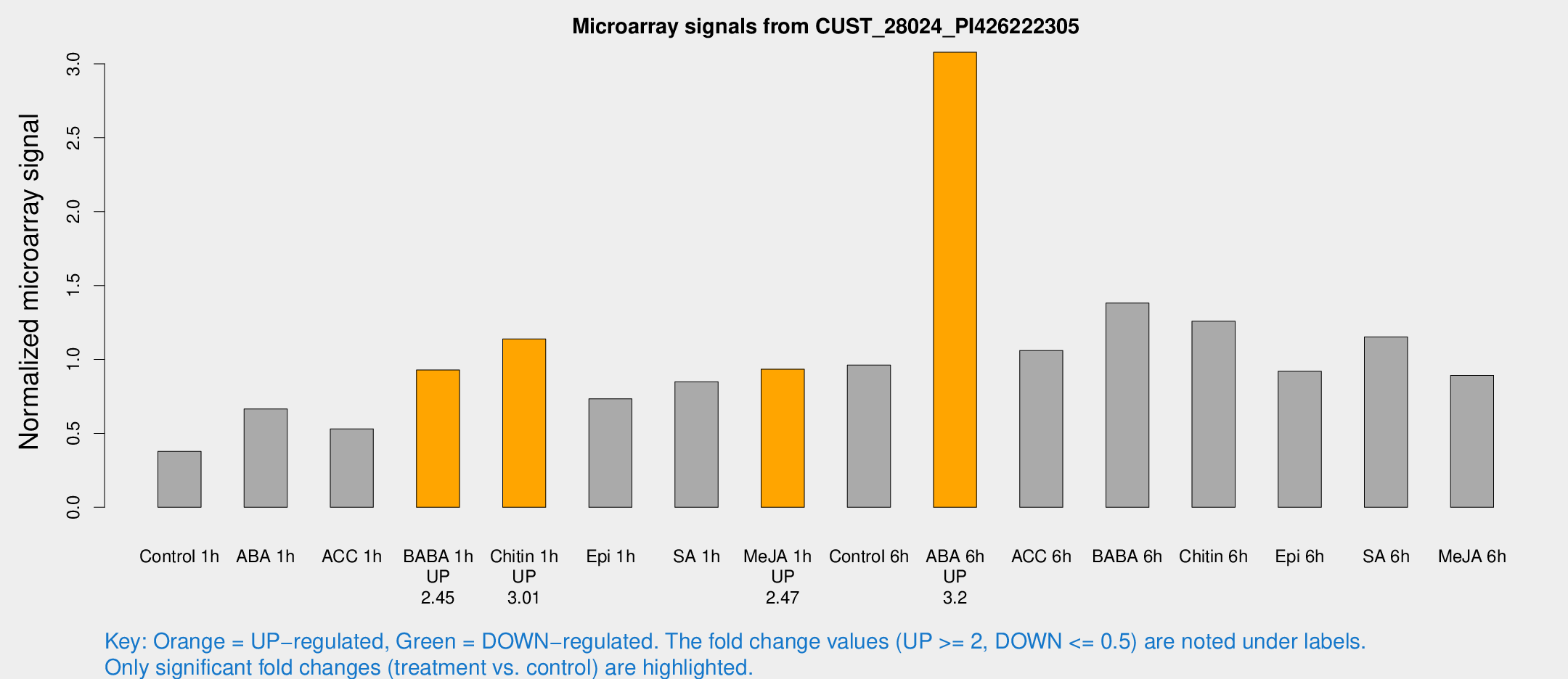
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1832.88 | 105.976 | 0.378618 | 0.0218718 |
| ABA 1h | 2926.61 | 462.345 | 0.664764 | 0.0784052 |
| ACC 1h | 2677.03 | 329.911 | 0.530167 | 0.0327817 |
| BABA 1h | 4633.09 | 1074.94 | 0.928501 | 0.160645 |
| Chitin 1h | 5076.3 | 821.729 | 1.13862 | 0.12963 |
| Epi 1h | 3143.59 | 452.609 | 0.733609 | 0.107671 |
| SA 1h | 4525.81 | 1263.31 | 0.849012 | 0.181693 |
| Me-JA 1h | 3730.38 | 400.936 | 0.934488 | 0.053961 |
| Control 6h | 4826.1 | 830.051 | 0.961596 | 0.0954798 |
| ABA 6h | 16112.6 | 2223.1 | 3.07828 | 0.256344 |
| ACC 6h | 6030.63 | 973.059 | 1.06051 | 0.0612334 |
| BABA 6h | 7508.23 | 482.103 | 1.38127 | 0.0869558 |
| Chitin 6h | 6496.91 | 376.038 | 1.25886 | 0.0726853 |
| Epi 6h | 5026.88 | 290.596 | 0.92068 | 0.0531615 |
| SA 6h | 5562.41 | 520.192 | 1.15256 | 0.0774354 |
| Me-JA 6h | 4818.22 | 1430.35 | 0.892418 | 0.260556 |
Source Transcript PGSC0003DMT400081977 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G32930.1 | +1 | 4e-102 | 303 | 144/196 (73%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast envelope; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 13 growth stages; Has 57 Blast hits to 57 proteins in 11 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 57; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:13489595-13490704 FORWARD LENGTH=249 |
