Probe CUST_25589_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25589_PI426222305 | JHI_St_60k_v1 | DMT400037313 | TTTTGTTGTTTGGGAGCCACCAGCAGAGCAGAAGATTATCTGGGTTACATTTGTTTTTTG |
All Microarray Probes Designed to Gene DMG400014398
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25589_PI426222305 | JHI_St_60k_v1 | DMT400037313 | TTTTGTTGTTTGGGAGCCACCAGCAGAGCAGAAGATTATCTGGGTTACATTTGTTTTTTG |
| CUST_25671_PI426222305 | JHI_St_60k_v1 | DMT400037312 | TTTTGTTGTTTGGGAGCCACCAGCAGAGCAGAAGATTATCTGGGTTACATTTGTTTTTTG |
| CUST_25735_PI426222305 | JHI_St_60k_v1 | DMT400037311 | ATCTTGATATAGGCAAAACACTTGGTCGAATTCATAGAGGGATGTGGAGGGATCCTGGTG |
Microarray Signals from CUST_25589_PI426222305
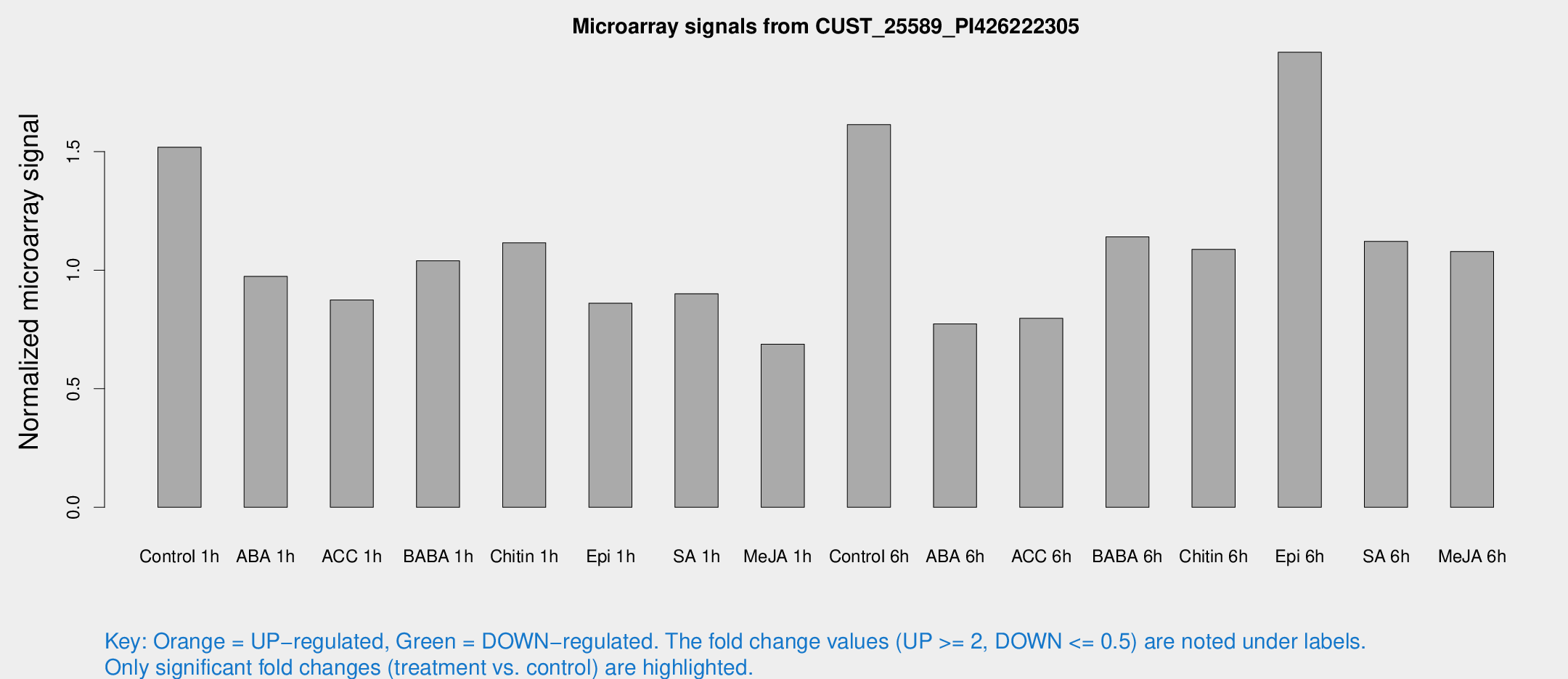
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 61.1265 | 5.86968 | 1.51843 | 0.113177 |
| ABA 1h | 34.6277 | 3.38518 | 0.97344 | 0.0968362 |
| ACC 1h | 36.4799 | 4.82349 | 0.87422 | 0.098409 |
| BABA 1h | 40.9403 | 6.35094 | 1.03937 | 0.101677 |
| Chitin 1h | 40.5263 | 4.63072 | 1.11527 | 0.102114 |
| Epi 1h | 31.0719 | 6.82696 | 0.860456 | 0.203236 |
| SA 1h | 38.2701 | 6.80929 | 0.900338 | 0.135634 |
| Me-JA 1h | 23.9268 | 6.41749 | 0.687424 | 0.239258 |
| Control 6h | 68.9627 | 18.2064 | 1.61332 | 0.626628 |
| ABA 6h | 35.3221 | 9.39582 | 0.773382 | 0.205969 |
| ACC 6h | 36.7467 | 4.13352 | 0.796689 | 0.0887019 |
| BABA 6h | 52.2534 | 8.76102 | 1.14013 | 0.145218 |
| Chitin 6h | 46.8722 | 5.84848 | 1.08793 | 0.158363 |
| Epi 6h | 141.725 | 98.0844 | 1.91874 | 1.89427 |
| SA 6h | 47.9516 | 12.4805 | 1.12156 | 0.500028 |
| Me-JA 6h | 44.3918 | 8.05914 | 1.07867 | 0.156495 |
Source Transcript PGSC0003DMT400037313 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G54780.1 | +3 | 0.0 | 633 | 314/444 (71%) | Ypt/Rab-GAP domain of gyp1p superfamily protein | chr5:22248696-22251692 REVERSE LENGTH=432 |
