Probe CUST_24832_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24832_PI426222305 | JHI_St_60k_v1 | DMT400024052 | CATTTGTTGCAAGGAAGGGATAATTTGTAGTACAGGGAATTGGTGAAACTCTTATTGAAG |
All Microarray Probes Designed to Gene DMG400009304
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24832_PI426222305 | JHI_St_60k_v1 | DMT400024052 | CATTTGTTGCAAGGAAGGGATAATTTGTAGTACAGGGAATTGGTGAAACTCTTATTGAAG |
| CUST_24901_PI426222305 | JHI_St_60k_v1 | DMT400024050 | CATTTGTTGCAAGGAAGGGATAATTTGTAGTACAGGGAATTGGTGAAACTCTTATTGAAG |
| CUST_24807_PI426222305 | JHI_St_60k_v1 | DMT400024051 | TACACGAGCAGCTTGAGGTACACATTAAACCTTTTCTCTAGAACAATTCCTAGTCCTAAC |
| CUST_24954_PI426222305 | JHI_St_60k_v1 | DMT400024053 | GCGCAAGTTGGCTTGGGATTGATGTAATGTAATTGTTTCTTTTAATGGGTGATAAACTTT |
Microarray Signals from CUST_24832_PI426222305
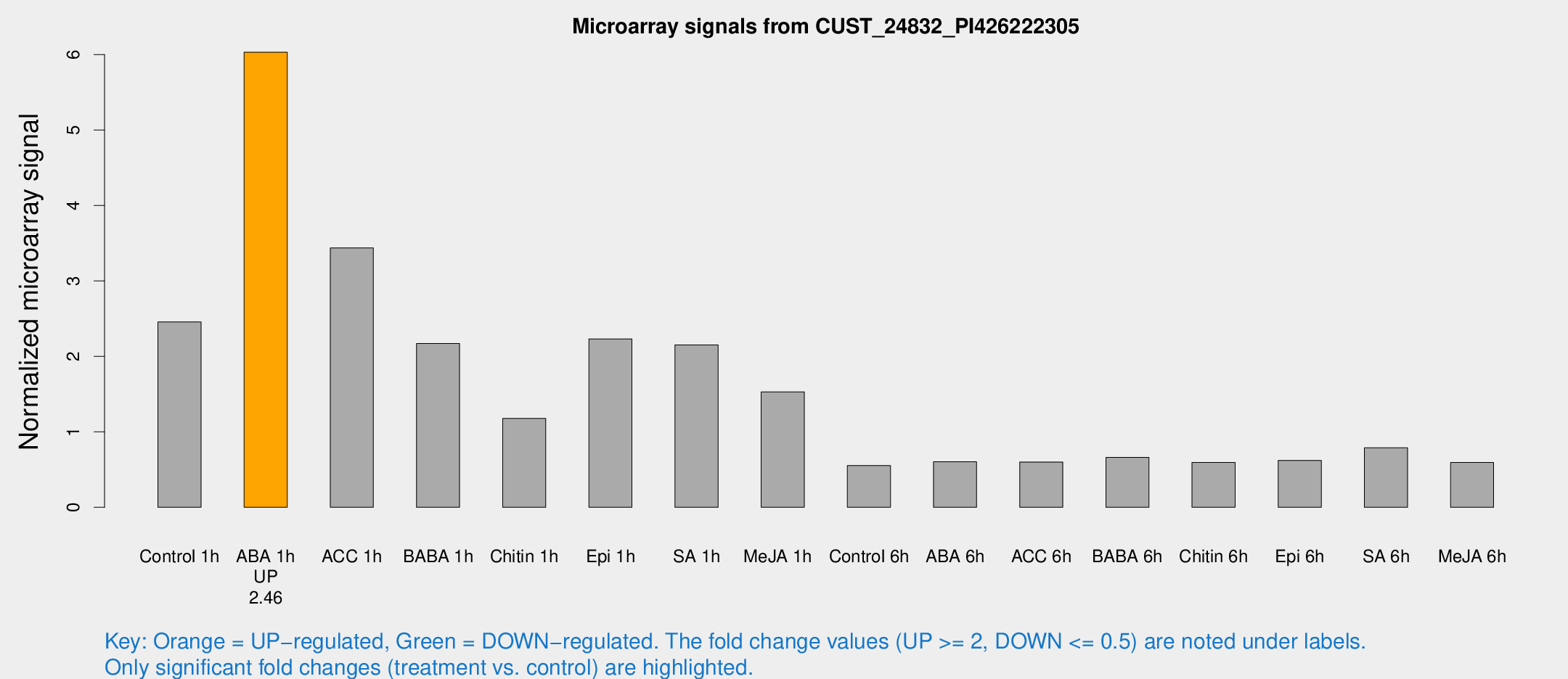
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 28.2597 | 4.08846 | 2.45676 | 0.359979 |
| ABA 1h | 60.9461 | 4.96311 | 6.03176 | 0.491255 |
| ACC 1h | 41.0056 | 5.41635 | 3.43887 | 0.462658 |
| BABA 1h | 26.3294 | 8.62682 | 2.17064 | 1.02129 |
| Chitin 1h | 13.5254 | 4.8127 | 1.17752 | 0.401154 |
| Epi 1h | 23.0804 | 5.22299 | 2.23231 | 0.539376 |
| SA 1h | 25.9061 | 4.00378 | 2.1532 | 0.501645 |
| Me-JA 1h | 14.2644 | 3.61307 | 1.53011 | 0.391221 |
| Control 6h | 6.32978 | 3.67245 | 0.554132 | 0.321486 |
| ABA 6h | 7.5431 | 3.78616 | 0.604412 | 0.315128 |
| ACC 6h | 8.14153 | 4.46416 | 0.599946 | 0.329843 |
| BABA 6h | 8.89622 | 4.05218 | 0.661314 | 0.329071 |
| Chitin 6h | 7.23887 | 4.2048 | 0.595087 | 0.344709 |
| Epi 6h | 8.02542 | 4.52797 | 0.621657 | 0.346684 |
| SA 6h | 9.57012 | 4.04659 | 0.788882 | 0.382594 |
| Me-JA 6h | 6.79344 | 3.65418 | 0.593394 | 0.322359 |
Source Transcript PGSC0003DMT400024052 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G45580.1 | +1 | 5e-77 | 243 | 141/264 (53%) | Homeodomain-like superfamily protein | chr5:18481092-18482598 REVERSE LENGTH=264 |
