Probe CUST_24460_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24460_PI426222305 | JHI_St_60k_v1 | DMT400035752 | GGAGAGGTCATTATTGGTGGACTTGTTACTGCTATTTTTGCTGCTGTTTATTGTTATATT |
All Microarray Probes Designed to Gene DMG400013754
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24460_PI426222305 | JHI_St_60k_v1 | DMT400035752 | GGAGAGGTCATTATTGGTGGACTTGTTACTGCTATTTTTGCTGCTGTTTATTGTTATATT |
Microarray Signals from CUST_24460_PI426222305
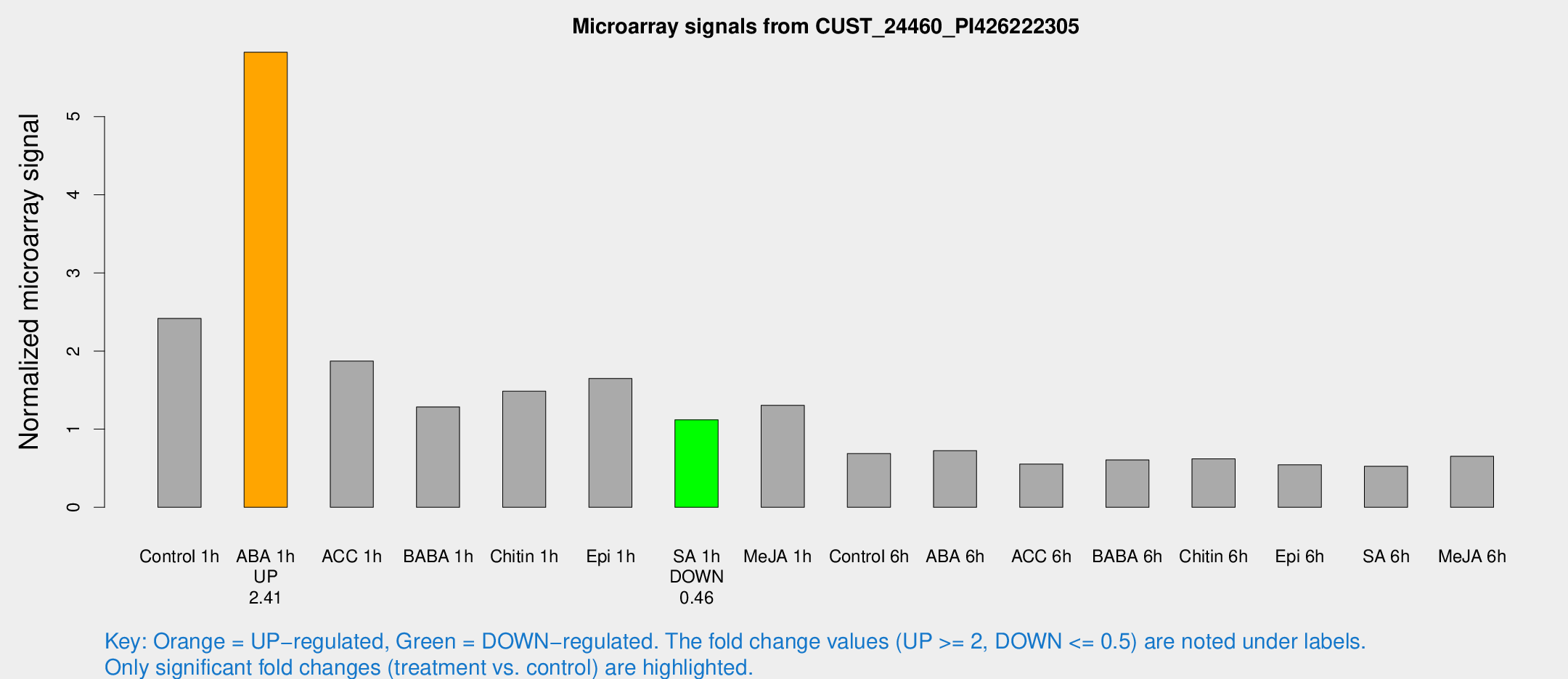
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 970.881 | 116.941 | 2.41726 | 0.168668 |
| ABA 1h | 2056.73 | 208.091 | 5.82337 | 0.643634 |
| ACC 1h | 778.238 | 115.092 | 1.87132 | 0.167115 |
| BABA 1h | 493.467 | 44.7129 | 1.28446 | 0.0840378 |
| Chitin 1h | 541.596 | 80.8172 | 1.48588 | 0.137529 |
| Epi 1h | 579.156 | 92.2093 | 1.64887 | 0.232814 |
| SA 1h | 458.594 | 39.8273 | 1.1203 | 0.0722008 |
| Me-JA 1h | 434.289 | 74.3692 | 1.30517 | 0.209225 |
| Control 6h | 286.278 | 58.1253 | 0.688734 | 0.0908889 |
| ABA 6h | 311.192 | 46.2229 | 0.725155 | 0.138336 |
| ACC 6h | 254.84 | 32.1947 | 0.553439 | 0.0332168 |
| BABA 6h | 276.605 | 50.8989 | 0.605423 | 0.0865027 |
| Chitin 6h | 263.579 | 24.5149 | 0.62089 | 0.0524339 |
| Epi 6h | 243.495 | 15.0961 | 0.544086 | 0.0455524 |
| SA 6h | 209.449 | 29.771 | 0.525403 | 0.0845055 |
| Me-JA 6h | 259.931 | 29.5338 | 0.652106 | 0.0386944 |
Source Transcript PGSC0003DMT400035752 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G09280.1 | +1 | 2e-11 | 58 | 37/87 (43%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: endomembrane system; EXPRESSED IN: root; Has 31 Blast hits to 31 proteins in 9 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 31; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:2850920-2851258 REVERSE LENGTH=112 |
