Probe CUST_2381_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2381_PI426222305 | JHI_St_60k_v1 | DMT400072432 | TGGTGCTTTTGTTGGTGGTAATATAACACTTTTACGTGGAATTTTGTATTGGATTGCCCA |
All Microarray Probes Designed to Gene DMG400028182
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2381_PI426222305 | JHI_St_60k_v1 | DMT400072432 | TGGTGCTTTTGTTGGTGGTAATATAACACTTTTACGTGGAATTTTGTATTGGATTGCCCA |
Microarray Signals from CUST_2381_PI426222305
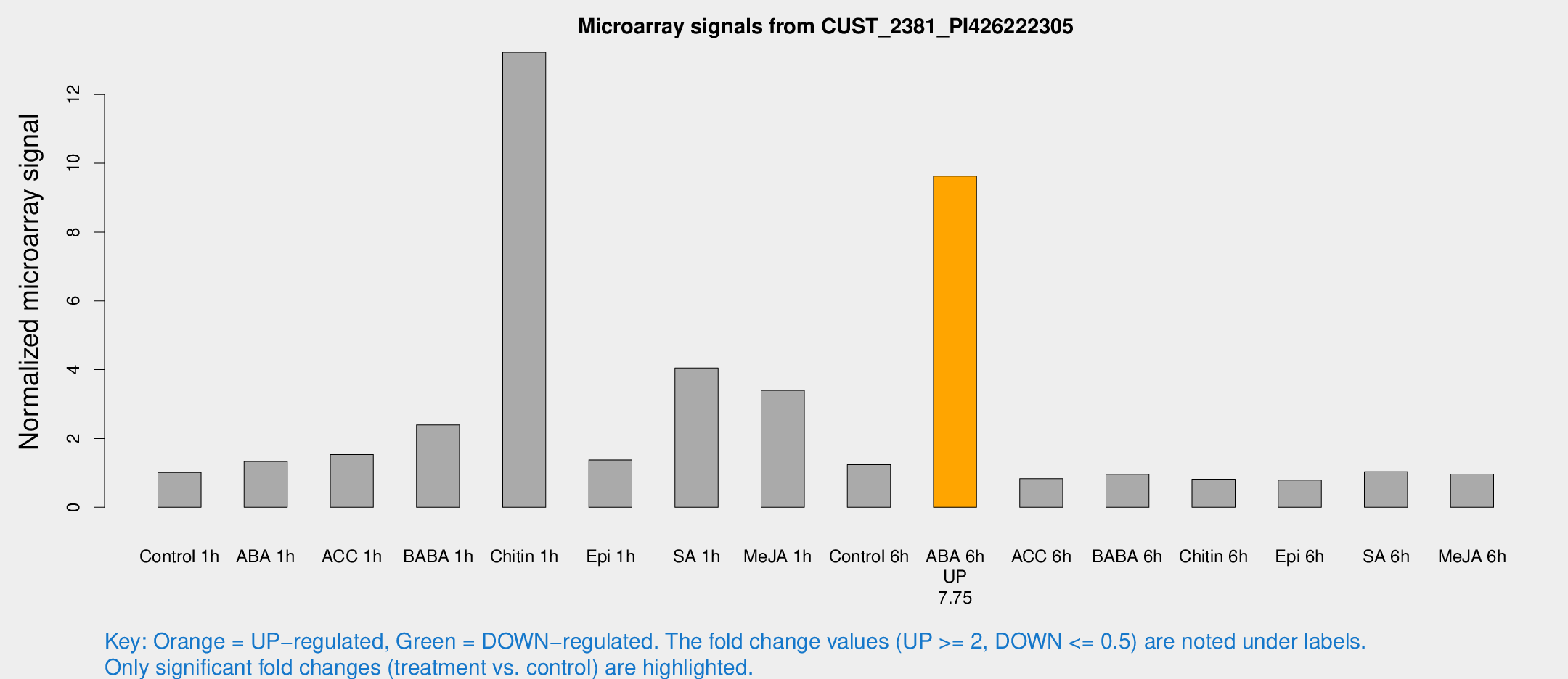
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.85672 | 2.85075 | 1.01135 | 0.472679 |
| ABA 1h | 8.89664 | 4.27383 | 1.3333 | 0.573892 |
| ACC 1h | 16.9242 | 11.8461 | 1.53803 | 1.98484 |
| BABA 1h | 19.0678 | 7.80314 | 2.39274 | 1.26501 |
| Chitin 1h | 90.6489 | 39.1899 | 13.2276 | 6.16061 |
| Epi 1h | 9.15622 | 4.45504 | 1.3786 | 0.717434 |
| SA 1h | 37.2712 | 19.3398 | 4.04759 | 3.65156 |
| Me-JA 1h | 23.5575 | 12.7596 | 3.40149 | 2.21808 |
| Control 6h | 10.0121 | 5.00827 | 1.24166 | 0.574764 |
| ABA 6h | 70.8228 | 19.4502 | 9.62807 | 2.36399 |
| ACC 6h | 6.24394 | 3.56216 | 0.833631 | 0.464076 |
| BABA 6h | 7.14654 | 3.30821 | 0.958808 | 0.477791 |
| Chitin 6h | 5.60549 | 3.25329 | 0.818819 | 0.475141 |
| Epi 6h | 5.80491 | 3.39286 | 0.794749 | 0.461074 |
| SA 6h | 6.88699 | 3.12473 | 1.03302 | 0.512578 |
| Me-JA 6h | 6.34528 | 2.93299 | 0.966427 | 0.474137 |
Source Transcript PGSC0003DMT400072432 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g083880.1 | +1 | 1e-161 | 458 | 243/249 (98%) | evidence_code:10F0H1E1IEG genomic_reference:SL2.50ch10 gene_region:62931204-62932616 transcript_region:SL2.50ch10:62931204..62932616+ go_terms:GO:0016020,GO:0016021 functional_description:Aquaporin (AHRD V1 ***- D6BRE1_9ROSI); contains Interpro domain(s) IPR012269 Aquaporin |
| TAIR PP10 | AT2G36830.1 | +1 | 2e-138 | 399 | 210/250 (84%) | gamma tonoplast intrinsic protein | chr2:15445490-15446336 FORWARD LENGTH=251 |
