Probe CUST_23504_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23504_PI426222305 | JHI_St_60k_v1 | DMT400023917 | CGTAACAAGAACAACAGCTTAAGATTTCAAAAGCACCTCTTTCTCTGTAACTACAAACAT |
All Microarray Probes Designed to Gene DMG400009245
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23574_PI426222305 | JHI_St_60k_v1 | DMT400023915 | GTTCCTATTATAGCTGAAATTGATCTCTACAAGTATAATCCATGGGATCTACCTGGTAAT |
| CUST_23504_PI426222305 | JHI_St_60k_v1 | DMT400023917 | CGTAACAAGAACAACAGCTTAAGATTTCAAAAGCACCTCTTTCTCTGTAACTACAAACAT |
| CUST_23512_PI426222305 | JHI_St_60k_v1 | DMT400023916 | TTGCTGTTCCTATTATAGCTGAAATTGATCTCTACAAGTATAATCCATGGGATCTACCTG |
Microarray Signals from CUST_23504_PI426222305
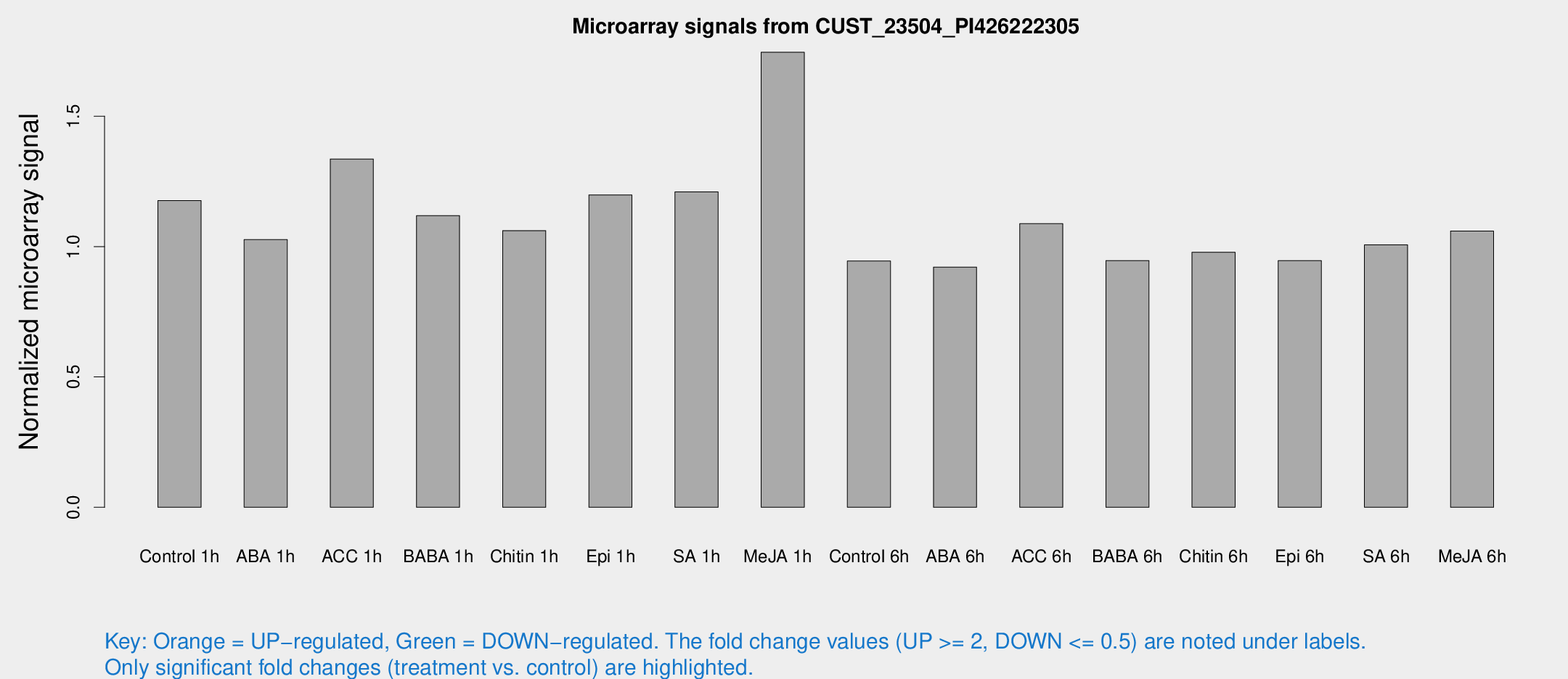
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.92229 | 2.92031 | 1.17646 | 0.556118 |
| ABA 1h | 5.00898 | 2.82903 | 1.02679 | 0.573264 |
| ACC 1h | 8.49248 | 3.22064 | 1.33604 | 0.629043 |
| BABA 1h | 6.19943 | 3.08897 | 1.11877 | 0.582397 |
| Chitin 1h | 5.14793 | 2.94905 | 1.06127 | 0.582901 |
| Epi 1h | 5.69788 | 2.91834 | 1.19817 | 0.606623 |
| SA 1h | 7.73455 | 2.95639 | 1.21 | 0.568019 |
| Me-JA 1h | 8.32328 | 3.03987 | 1.74537 | 0.70152 |
| Control 6h | 5.28183 | 3.05959 | 0.94465 | 0.547042 |
| ABA 6h | 5.46774 | 3.16918 | 0.921463 | 0.533696 |
| ACC 6h | 7.09236 | 3.72083 | 1.08799 | 0.562967 |
| BABA 6h | 5.92347 | 3.44106 | 0.946445 | 0.548333 |
| Chitin 6h | 5.80915 | 3.36472 | 0.9782 | 0.566402 |
| Epi 6h | 6.00081 | 3.50804 | 0.946328 | 0.548569 |
| SA 6h | 5.55432 | 3.21795 | 1.00636 | 0.582764 |
| Me-JA 6h | 5.98096 | 3.0771 | 1.05986 | 0.559424 |
Source Transcript PGSC0003DMT400023917 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G01720.1 | +3 | 1e-134 | 392 | 205/301 (68%) | NAC (No Apical Meristem) domain transcriptional regulator superfamily protein | chr1:268471-269514 FORWARD LENGTH=289 |
