Probe CUST_22745_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22745_PI426222305 | JHI_St_60k_v1 | DMT400078225 | TCGCACCTCTTCAATCCAGATTGTTGGCTATAAATTGTAAGCTACAAATAAATATGGTGT |
All Microarray Probes Designed to Gene DMG400030440
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22745_PI426222305 | JHI_St_60k_v1 | DMT400078225 | TCGCACCTCTTCAATCCAGATTGTTGGCTATAAATTGTAAGCTACAAATAAATATGGTGT |
Microarray Signals from CUST_22745_PI426222305
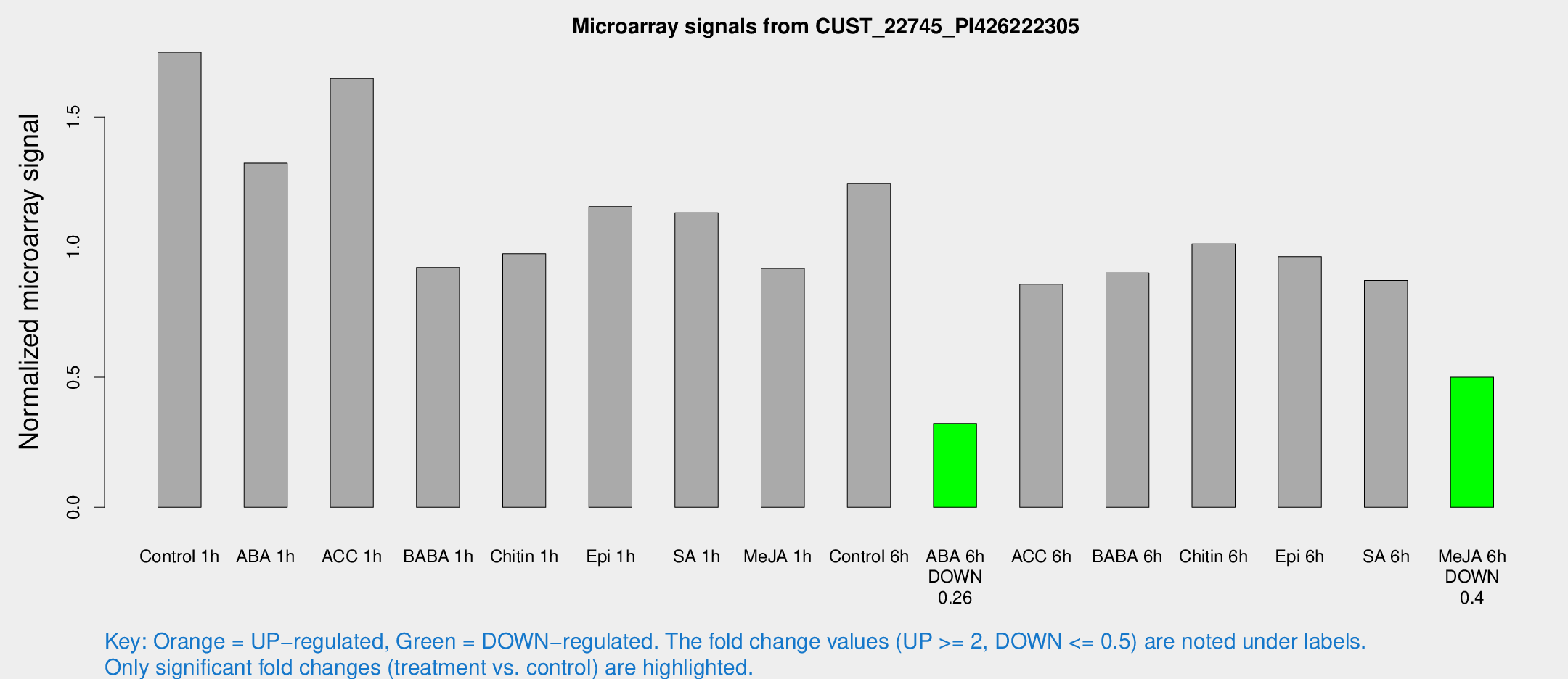
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1945.6 | 245.962 | 1.74855 | 0.102314 |
| ABA 1h | 1300.05 | 160.343 | 1.32241 | 0.243299 |
| ACC 1h | 1859.57 | 107.766 | 1.64719 | 0.223503 |
| BABA 1h | 1018.71 | 202.046 | 0.921359 | 0.110818 |
| Chitin 1h | 989.676 | 180.678 | 0.974249 | 0.190462 |
| Epi 1h | 1097.04 | 63.4845 | 1.15532 | 0.0667696 |
| SA 1h | 1280.72 | 95.0164 | 1.13172 | 0.0804486 |
| Me-JA 1h | 825.964 | 63.1827 | 0.918141 | 0.0859233 |
| Control 6h | 1414.81 | 264.878 | 1.2446 | 0.168045 |
| ABA 6h | 375.379 | 21.9183 | 0.321832 | 0.0299033 |
| ACC 6h | 1108.71 | 187.945 | 0.857015 | 0.0529137 |
| BABA 6h | 1145.83 | 221.997 | 0.900176 | 0.191208 |
| Chitin 6h | 1197.09 | 145.061 | 1.01234 | 0.0922223 |
| Epi 6h | 1192.88 | 69.0975 | 0.963572 | 0.11431 |
| SA 6h | 974.318 | 159.127 | 0.871998 | 0.0542961 |
| Me-JA 6h | 584.045 | 157.756 | 0.499544 | 0.0921022 |
Source Transcript PGSC0003DMT400078225 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G47070.1 | +2 | 3e-07 | 48 | 34/66 (52%) | LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 13 growth stages; CONTAINS InterPro DOMAIN/s: Thylakoid soluble phosphoprotein TSP9 (InterPro:IPR021584); Has 37 Blast hits to 37 proteins in 10 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 37; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:17337205-17337507 REVERSE LENGTH=100 |
