Probe CUST_22420_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22420_PI426222305 | JHI_St_60k_v1 | DMT400039510 | CTTGGCTTTGTCGTGAATTTCATTTACATATAGAACCGTTGAGAAGGCAGCAAGATTTAC |
All Microarray Probes Designed to Gene DMG400015277
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22420_PI426222305 | JHI_St_60k_v1 | DMT400039510 | CTTGGCTTTGTCGTGAATTTCATTTACATATAGAACCGTTGAGAAGGCAGCAAGATTTAC |
| CUST_22399_PI426222305 | JHI_St_60k_v1 | DMT400039512 | GGAGCATTCATTATATAATGTCTTGGTATGCTGCCAAAATTCATTGGATCCATCAAATAG |
| CUST_22503_PI426222305 | JHI_St_60k_v1 | DMT400039513 | GGAGCATTCATTATATAATGTCTTGGTATGCTGCCAAAATTCATTGGATCCATCAAATAG |
Microarray Signals from CUST_22420_PI426222305
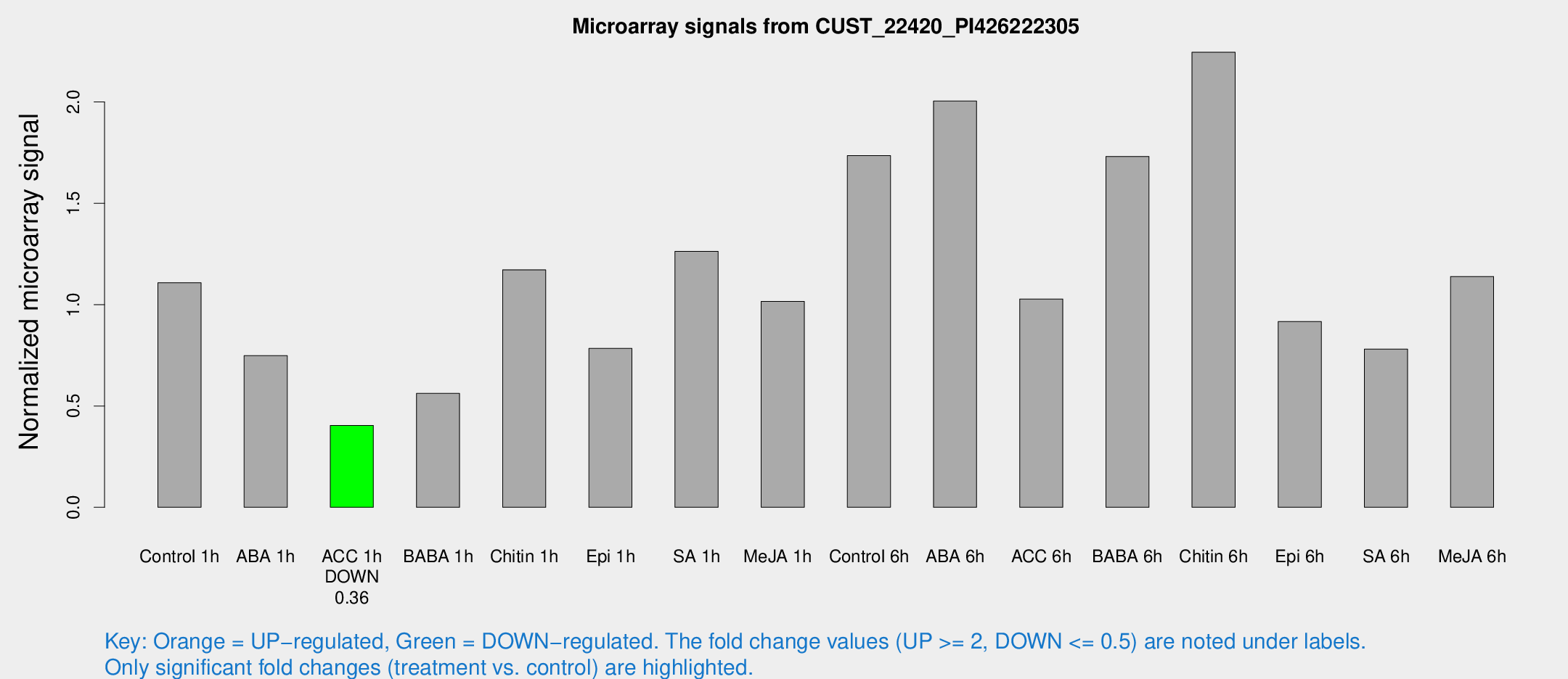
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.6884 | 7.65653 | 1.10813 | 0.249329 |
| ABA 1h | 14.5557 | 3.59288 | 0.748076 | 0.199387 |
| ACC 1h | 8.95294 | 3.97225 | 0.403886 | 0.185208 |
| BABA 1h | 13.0284 | 4.49259 | 0.561881 | 0.223053 |
| Chitin 1h | 23.3548 | 4.29969 | 1.17167 | 0.210782 |
| Epi 1h | 15.2305 | 3.70852 | 0.783797 | 0.211925 |
| SA 1h | 28.3521 | 4.54853 | 1.26239 | 0.27714 |
| Me-JA 1h | 18.3592 | 3.76461 | 1.0156 | 0.30088 |
| Control 6h | 38.5542 | 7.67094 | 1.73493 | 0.260865 |
| ABA 6h | 88.2813 | 65.4239 | 2.00446 | 2.93422 |
| ACC 6h | 25.5478 | 4.91489 | 1.02778 | 0.195344 |
| BABA 6h | 51.663 | 21.9119 | 1.73123 | 0.935471 |
| Chitin 6h | 64.2044 | 31.5033 | 2.2453 | 1.22962 |
| Epi 6h | 28.8631 | 14.3093 | 0.916905 | 0.660824 |
| SA 6h | 19.1129 | 6.06016 | 0.780875 | 0.472201 |
| Me-JA 6h | 26.9495 | 8.09844 | 1.13886 | 0.34573 |
Source Transcript PGSC0003DMT400039510 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73350.2 | +1 | 6e-24 | 96 | 76/92 (83%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; EXPRESSED IN: 25 plant structures; EXPRESSED DURING: 15 growth stages; Has 50 Blast hits to 50 proteins in 20 species: Archae - 0; Bacteria - 0; Metazoa - 7; Fungi - 3; Plants - 36; Viruses - 0; Other Eukaryotes - 4 (source: NCBI BLink). | chr1:27575683-27577126 REVERSE LENGTH=210 |
