Probe CUST_20854_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20854_PI426222305 | JHI_St_60k_v1 | DMT400011968 | TCATTTATATGGCGTTCTGGTGGCCGTTGATTGTTATCGCTATCGCATTTGCGATCTGTA |
All Microarray Probes Designed to Gene DMG400004698
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20836_PI426222305 | JHI_St_60k_v1 | DMT400011970 | TCCTTCAAATTCCTTCTAAGAGTTGTGTGAAAAACCCTAATTAGTCTTCTTCGAAGCTTC |
| CUST_20745_PI426222305 | JHI_St_60k_v1 | DMT400011969 | ACTTGTATATTGTCACACTTAATTTTGCTCATTTAGATTCCCTCGAGAAGGCATACAGAC |
| CUST_20854_PI426222305 | JHI_St_60k_v1 | DMT400011968 | TCATTTATATGGCGTTCTGGTGGCCGTTGATTGTTATCGCTATCGCATTTGCGATCTGTA |
Microarray Signals from CUST_20854_PI426222305
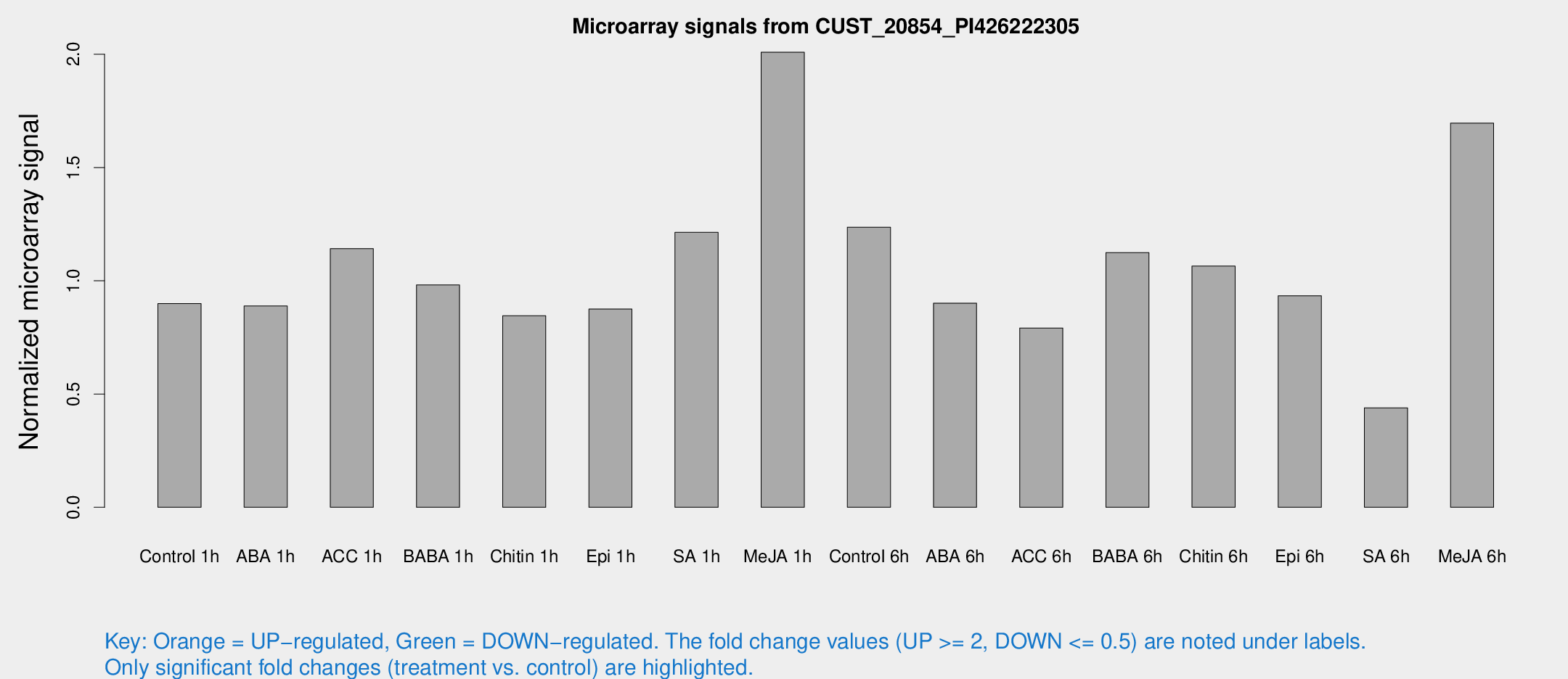
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 59.116 | 8.56669 | 0.899399 | 0.106839 |
| ABA 1h | 51.5555 | 7.43578 | 0.888809 | 0.106009 |
| ACC 1h | 79.1103 | 16.1765 | 1.14192 | 0.174917 |
| BABA 1h | 61.1585 | 4.97096 | 0.981684 | 0.0807387 |
| Chitin 1h | 50.673 | 9.28497 | 0.84553 | 0.13438 |
| Epi 1h | 49.1969 | 5.10334 | 0.87497 | 0.0820547 |
| SA 1h | 83.3638 | 17.085 | 1.21411 | 0.280032 |
| Me-JA 1h | 107.896 | 17.3802 | 2.00881 | 0.344996 |
| Control 6h | 88.5213 | 26.6144 | 1.23662 | 0.317667 |
| ABA 6h | 62.8593 | 8.67342 | 0.901081 | 0.0959936 |
| ACC 6h | 60.4941 | 12.0675 | 0.791485 | 0.0729335 |
| BABA 6h | 81.1753 | 6.18465 | 1.12433 | 0.0855293 |
| Chitin 6h | 73.1307 | 5.76379 | 1.0646 | 0.0841268 |
| Epi 6h | 71.5813 | 17.0816 | 0.933926 | 0.269933 |
| SA 6h | 30.8897 | 10.299 | 0.439266 | 0.137352 |
| Me-JA 6h | 117.989 | 32.2425 | 1.69607 | 0.373956 |
Source Transcript PGSC0003DMT400011968 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G66658.2 | +1 | 0.0 | 706 | 343/434 (79%) | aldehyde dehydrogenase 22A1 | chr3:2095341-2099013 REVERSE LENGTH=596 |
