Probe CUST_2084_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2084_PI426222305 | JHI_St_60k_v1 | DMT400072673 | TCTCCAGCCAAATGTATGTTTCCCCCTATTTTTGCCTTCTTATGAATGTGTTGCACCCCA |
All Microarray Probes Designed to Gene DMG400028278
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2084_PI426222305 | JHI_St_60k_v1 | DMT400072673 | TCTCCAGCCAAATGTATGTTTCCCCCTATTTTTGCCTTCTTATGAATGTGTTGCACCCCA |
Microarray Signals from CUST_2084_PI426222305
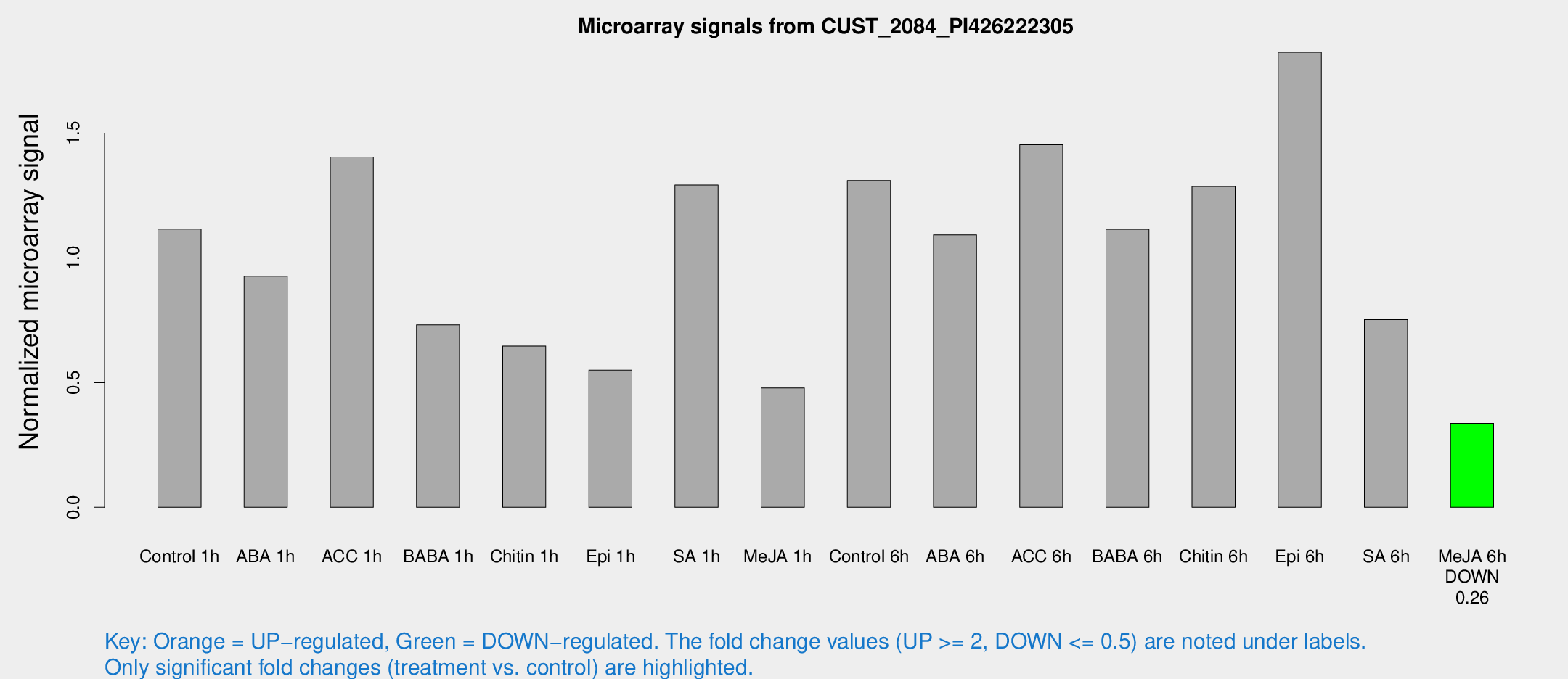
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 23.6953 | 6.04244 | 1.11596 | 0.296201 |
| ABA 1h | 18.8003 | 7.05382 | 0.926916 | 0.340232 |
| ACC 1h | 36.4303 | 13.7207 | 1.40454 | 0.840254 |
| BABA 1h | 19.3295 | 11.4001 | 0.731462 | 0.454161 |
| Chitin 1h | 11.7474 | 2.91689 | 0.646938 | 0.167149 |
| Epi 1h | 10.9015 | 4.34762 | 0.54991 | 0.241738 |
| SA 1h | 27.3289 | 5.61458 | 1.29215 | 0.221546 |
| Me-JA 1h | 7.75883 | 2.93669 | 0.479051 | 0.184487 |
| Control 6h | 26.6196 | 4.66839 | 1.3105 | 0.176518 |
| ABA 6h | 24.2958 | 5.91002 | 1.0927 | 0.265986 |
| ACC 6h | 36.57 | 11.6787 | 1.45355 | 0.392827 |
| BABA 6h | 25.2547 | 4.29407 | 1.1146 | 0.169295 |
| Chitin 6h | 27.0207 | 3.60921 | 1.28686 | 0.174733 |
| Epi 6h | 45.0061 | 15.4371 | 1.82461 | 0.677964 |
| SA 6h | 14.8386 | 3.2277 | 0.753129 | 0.1693 |
| Me-JA 6h | 6.70744 | 2.95658 | 0.337077 | 0.154879 |
Source Transcript PGSC0003DMT400072673 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g083860.1 | +2 | 0.0 | 890 | 466/485 (96%) | evidence_code:10F0H1E1IEG genomic_reference:SL2.50ch10 gene_region:62918325-62919809 transcript_region:SL2.50ch10:62918325..62919809- go_terms:GO:0008152 functional_description:UDP-glucosyltransferase family 1 protein (AHRD V1 ***- C6KI43_CITSI); contains Interpro domain(s) IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| TAIR PP10 | AT2G36790.1 | +2 | 1e-162 | 480 | 246/480 (51%) | UDP-glucosyl transferase 73C6 | chr2:15420339-15421826 REVERSE LENGTH=495 |
