Probe CUST_2072_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2072_PI426222305 | JHI_St_60k_v1 | DMT400027159 | CATTTCCTTCTTCTGTTAAAGCAGTACTTCCTCATCATCATCAAGAACTTCAAACAACAA |
All Microarray Probes Designed to Gene DMG400010478
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2072_PI426222305 | JHI_St_60k_v1 | DMT400027159 | CATTTCCTTCTTCTGTTAAAGCAGTACTTCCTCATCATCATCAAGAACTTCAAACAACAA |
Microarray Signals from CUST_2072_PI426222305
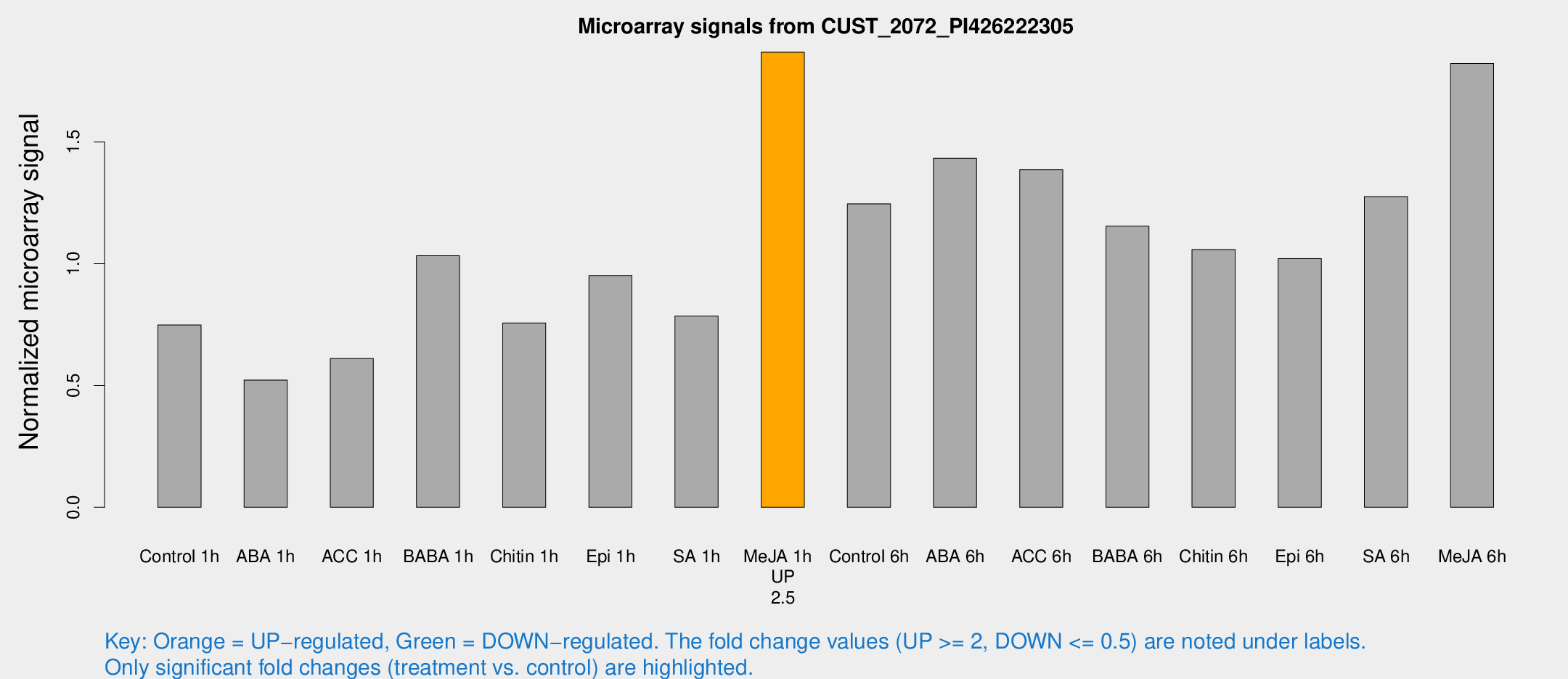
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 51.6599 | 4.15431 | 0.748447 | 0.0813481 |
| ABA 1h | 31.971 | 3.34088 | 0.522522 | 0.0736621 |
| ACC 1h | 45.1168 | 9.80028 | 0.610913 | 0.102892 |
| BABA 1h | 69.5843 | 8.80456 | 1.03273 | 0.0763229 |
| Chitin 1h | 48.0743 | 7.93541 | 0.756924 | 0.170043 |
| Epi 1h | 58.1597 | 9.2243 | 0.951968 | 0.156397 |
| SA 1h | 55.8959 | 5.30234 | 0.784826 | 0.0618888 |
| Me-JA 1h | 105.043 | 6.76419 | 1.86841 | 0.120071 |
| Control 6h | 91.915 | 22.6093 | 1.24597 | 0.245826 |
| ABA 6h | 113.266 | 28.3764 | 1.43252 | 0.373661 |
| ACC 6h | 116.265 | 25.9227 | 1.38639 | 0.30493 |
| BABA 6h | 94.0432 | 21.217 | 1.15433 | 0.260088 |
| Chitin 6h | 79.0389 | 10.9436 | 1.05867 | 0.154858 |
| Epi 6h | 79.1534 | 5.73423 | 1.02126 | 0.109586 |
| SA 6h | 104.78 | 40.7098 | 1.27541 | 0.927297 |
| Me-JA 6h | 130.214 | 27.6016 | 1.82219 | 0.233604 |
Source Transcript PGSC0003DMT400027159 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g084180.1 | +2 | 0.0 | 809 | 472/499 (95%) | evidence_code:10F0H1E1IEG genomic_reference:SL2.50ch10 gene_region:63159261-63161247 transcript_region:SL2.50ch10:63159261..63161247- go_terms:GO:0005515,GO:0003700 functional_description:C2H2L domain class transcription factor (AHRD V1 **-* D9ZIU4_MALDO); contains Interpro domain(s) IPR007087 Zinc finger, C2H2-type |
| TAIR PP10 | AT5G03150.1 | +2 | 2e-110 | 344 | 249/519 (48%) | C2H2-like zinc finger protein | chr5:745849-748678 FORWARD LENGTH=503 |
