Probe CUST_2033_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2033_PI426222305 | JHI_St_60k_v1 | DMT400072452 | CGCGCTGCGGAATTACAAAAGATTTTGGAACTCAAGATTGGATGAAATATCATATTACTT |
All Microarray Probes Designed to Gene DMG400028187
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2033_PI426222305 | JHI_St_60k_v1 | DMT400072452 | CGCGCTGCGGAATTACAAAAGATTTTGGAACTCAAGATTGGATGAAATATCATATTACTT |
Microarray Signals from CUST_2033_PI426222305
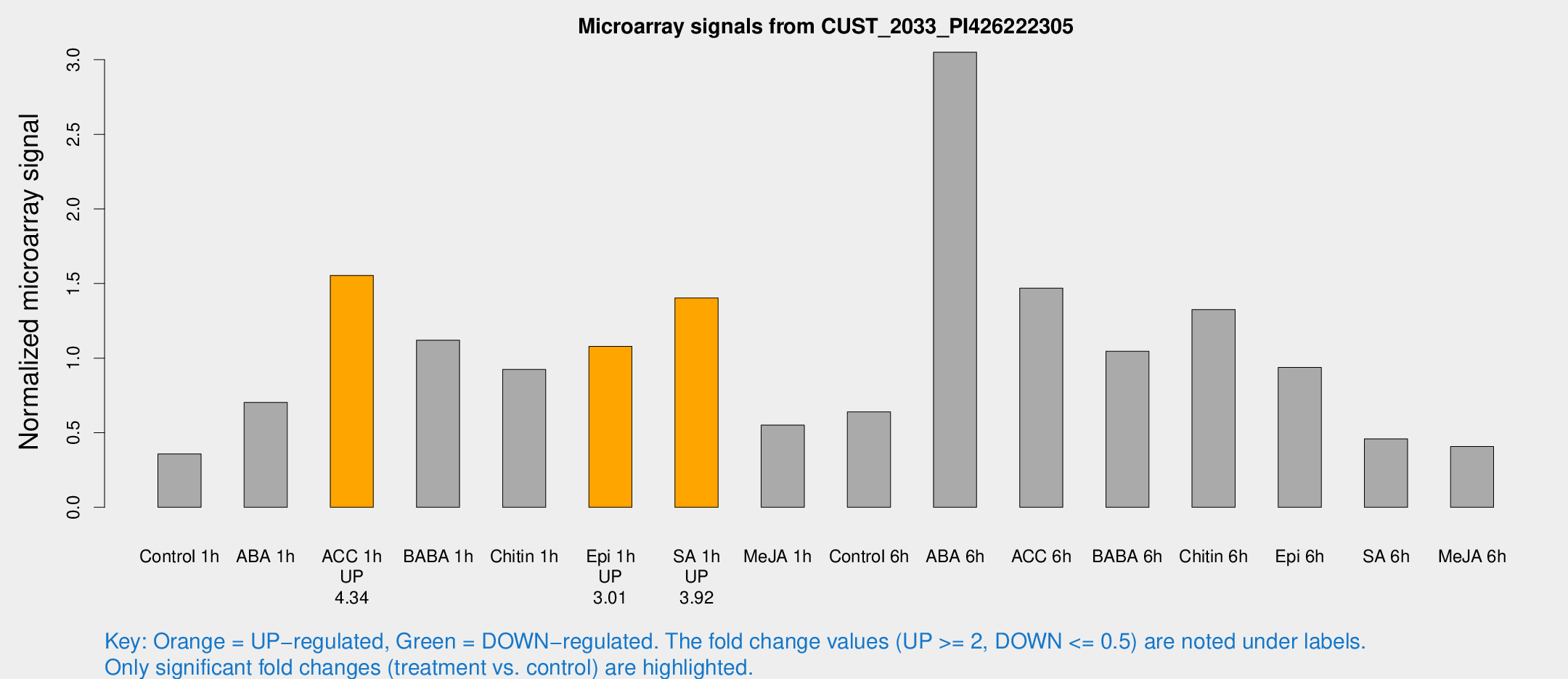
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.51716 | 3.20307 | 0.358329 | 0.207909 |
| ABA 1h | 10.5799 | 3.13279 | 0.703387 | 0.261765 |
| ACC 1h | 27.2718 | 7.78491 | 1.55453 | 0.431717 |
| BABA 1h | 17.6562 | 3.93481 | 1.11982 | 0.38025 |
| Chitin 1h | 14.4939 | 4.28027 | 0.924349 | 0.49541 |
| Epi 1h | 14.4101 | 3.27498 | 1.07835 | 0.24893 |
| SA 1h | 23.1879 | 5.08668 | 1.40294 | 0.225888 |
| Me-JA 1h | 7.158 | 3.29291 | 0.550464 | 0.272402 |
| Control 6h | 10.5021 | 3.41924 | 0.639517 | 0.24454 |
| ABA 6h | 73.5001 | 45.9664 | 3.05038 | 2.78651 |
| ACC 6h | 30.5694 | 12.7794 | 1.46918 | 0.400601 |
| BABA 6h | 24.0508 | 13.2818 | 1.04642 | 0.619715 |
| Chitin 6h | 22.2251 | 3.93061 | 1.32482 | 0.248101 |
| Epi 6h | 18.5264 | 5.79532 | 0.938065 | 0.478624 |
| SA 6h | 7.03407 | 3.5022 | 0.458685 | 0.231511 |
| Me-JA 6h | 6.32039 | 3.29096 | 0.407944 | 0.216735 |
Source Transcript PGSC0003DMT400072452 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g084030.1 | +3 | 2e-54 | 171 | 82/88 (93%) | evidence_code:10F0H0E0IEG genomic_reference:SL2.50ch10 gene_region:63026577-63027672 transcript_region:SL2.50ch10:63026577..63027672+ functional_description:Auxin responsive SAUR protein (AHRD V1 *-*- Q1SN26_MEDTR); contains Interpro domain(s) IPR003676 Auxin responsive SAUR protein |
| TAIR PP10 | AT2G28085.1 | +3 | 6e-19 | 79 | 39/71 (55%) | SAUR-like auxin-responsive protein family | chr2:11968182-11968556 REVERSE LENGTH=124 |
