Probe CUST_18783_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18783_PI426222305 | JHI_St_60k_v1 | DMT400001118 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
All Microarray Probes Designed to Gene DMG400000422
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18783_PI426222305 | JHI_St_60k_v1 | DMT400001118 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
| CUST_18866_PI426222305 | JHI_St_60k_v1 | DMT400001119 | ACCGTGTGTGGCATTAGATATGGTTGCTAGCATCACACAATCTCCAGCAGTTTCAAAAAA |
Microarray Signals from CUST_18783_PI426222305
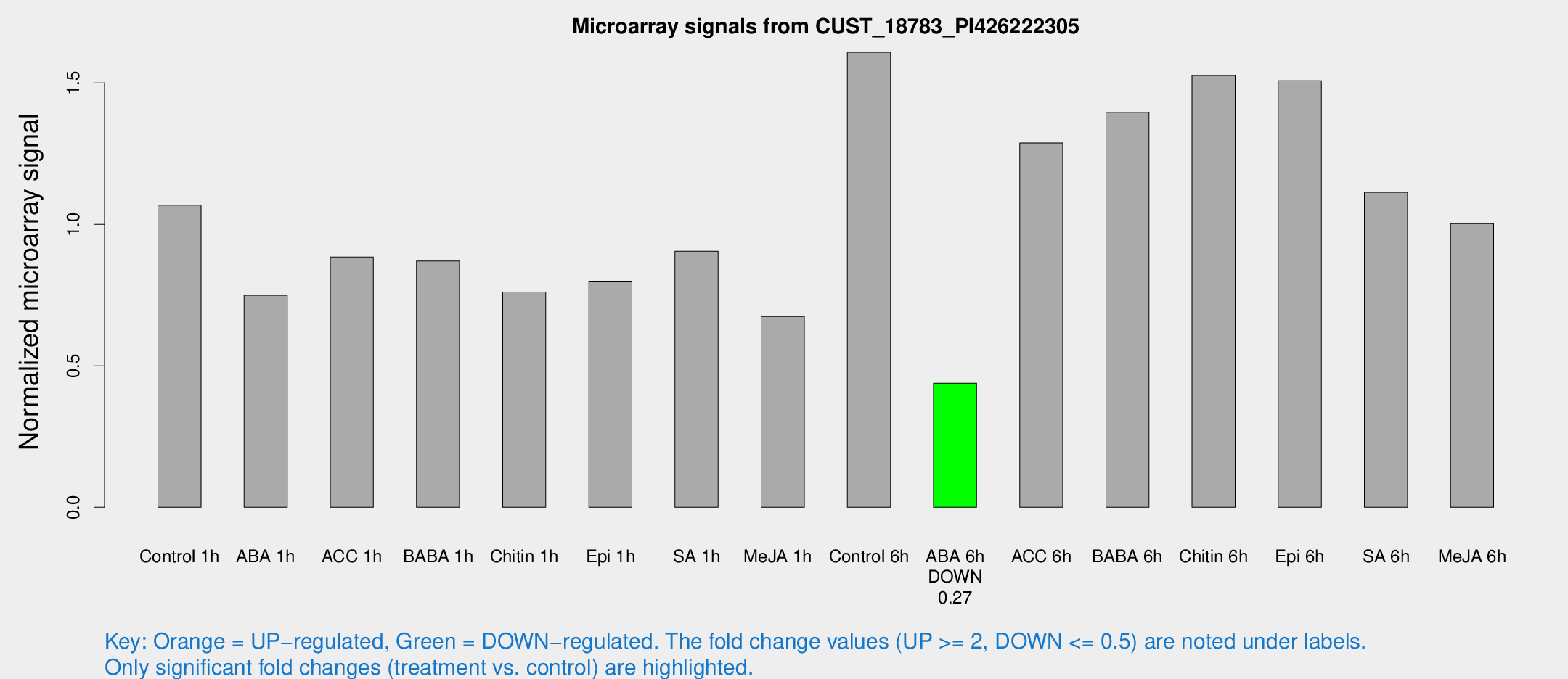
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2568.06 | 426.287 | 1.06793 | 0.109982 |
| ABA 1h | 1575.94 | 175.794 | 0.749654 | 0.100701 |
| ACC 1h | 2236.4 | 448.294 | 0.8846 | 0.136478 |
| BABA 1h | 2012.35 | 291.892 | 0.870575 | 0.0521273 |
| Chitin 1h | 1608 | 93.0558 | 0.761454 | 0.0541687 |
| Epi 1h | 1623.76 | 101.265 | 0.797002 | 0.046041 |
| SA 1h | 2195.7 | 187.954 | 0.905392 | 0.0971454 |
| Me-JA 1h | 1297.93 | 95.7671 | 0.674331 | 0.0389682 |
| Control 6h | 3968.6 | 804.873 | 1.6086 | 0.230993 |
| ABA 6h | 1096.4 | 63.5497 | 0.438782 | 0.0253696 |
| ACC 6h | 3507.05 | 386.308 | 1.28778 | 0.0849855 |
| BABA 6h | 3740.66 | 554.931 | 1.39658 | 0.195612 |
| Chitin 6h | 3809.18 | 219.966 | 1.52632 | 0.088134 |
| Epi 6h | 4037.26 | 460.53 | 1.50813 | 0.23048 |
| SA 6h | 2596.91 | 192.958 | 1.11393 | 0.13316 |
| Me-JA 6h | 2412.8 | 397.646 | 1.00252 | 0.0972645 |
Source Transcript PGSC0003DMT400001118 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G27920.1 | +1 | 0.0 | 563 | 281/413 (68%) | serine carboxypeptidase-like 51 | chr2:11885777-11889043 REVERSE LENGTH=461 |
