Probe CUST_18687_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18687_PI426222305 | JHI_St_60k_v1 | DMT400001179 | GGCTTCTTCACCTTCAACCTACAAGCCAATTGATGGAGAGGTTTAAATTTCTTTTATTTA |
All Microarray Probes Designed to Gene DMG400000443
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18687_PI426222305 | JHI_St_60k_v1 | DMT400001179 | GGCTTCTTCACCTTCAACCTACAAGCCAATTGATGGAGAGGTTTAAATTTCTTTTATTTA |
Microarray Signals from CUST_18687_PI426222305
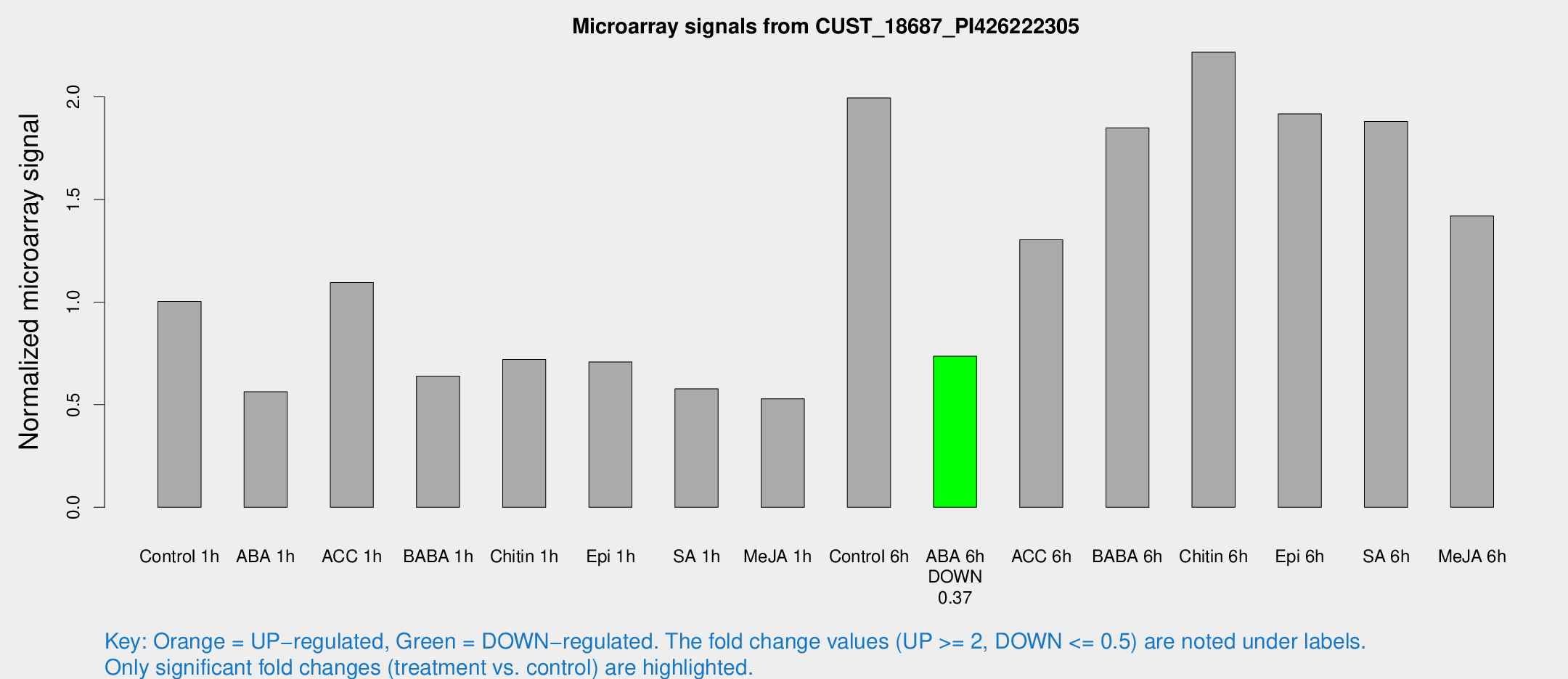
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1660.76 | 352.601 | 1.00277 | 0.15968 |
| ABA 1h | 793.205 | 77.498 | 0.56286 | 0.0332241 |
| ACC 1h | 1818.26 | 282.222 | 1.09515 | 0.100195 |
| BABA 1h | 1018.67 | 198.764 | 0.639321 | 0.075943 |
| Chitin 1h | 1042.96 | 146.635 | 0.720557 | 0.0936871 |
| Epi 1h | 969.712 | 56.1845 | 0.708437 | 0.0409724 |
| SA 1h | 942.029 | 72.2005 | 0.577617 | 0.0334143 |
| Me-JA 1h | 689.081 | 71.3501 | 0.528703 | 0.04396 |
| Control 6h | 3409.55 | 863.986 | 1.99479 | 0.423253 |
| ABA 6h | 1241.73 | 86.5482 | 0.736333 | 0.0425653 |
| ACC 6h | 2481.66 | 533.322 | 1.30377 | 0.163401 |
| BABA 6h | 3397.56 | 697.688 | 1.84852 | 0.343752 |
| Chitin 6h | 3736.81 | 216.416 | 2.21731 | 0.128036 |
| Epi 6h | 3458.62 | 394.305 | 1.91692 | 0.336039 |
| SA 6h | 2940.44 | 170.077 | 1.8801 | 0.304476 |
| Me-JA 6h | 2440.6 | 659.967 | 1.41972 | 0.338922 |
Source Transcript PGSC0003DMT400001179 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G59350.1 | +2 | 2e-28 | 115 | 97/259 (37%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: endomembrane system; EXPRESSED IN: 23 plant structures; EXPRESSED DURING: 13 growth stages; Has 1807 Blast hits to 1807 proteins in 277 species: Archae - 0; Bacteria - 0; Metazoa - 736; Fungi - 347; Plants - 385; Viruses - 0; Other Eukaryotes - 339 (source: NCBI BLink). | chr5:23941132-23941995 FORWARD LENGTH=287 |
