Probe CUST_17712_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17712_PI426222305 | JHI_St_60k_v1 | DMT400066804 | TTGTTGTGAAGTTCCACTTCTGAGAAACTCTTGAGCTCCATGACTTGATCCTCCTCTCTC |
All Microarray Probes Designed to Gene DMG400025975
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17688_PI426222305 | JHI_St_60k_v1 | DMT400066805 | TTGTTGTGAAGTTCCACTTCTGAGAAACTCTTGAGCTCCATGACTTGATCCTCCTCTCTC |
| CUST_17780_PI426222305 | JHI_St_60k_v1 | DMT400066806 | GAAGGGAGGGTATCCTTTGAACAGTCTGTTACAACTTGCAAGAAATTATTCTGAAGTGGC |
| CUST_17798_PI426222305 | JHI_St_60k_v1 | DMT400066802 | TTAACGGTAACCCAATTTCATCCGCAAAGCTCTCATACACCTACATTCATATACTGGAAC |
| CUST_17712_PI426222305 | JHI_St_60k_v1 | DMT400066804 | TTGTTGTGAAGTTCCACTTCTGAGAAACTCTTGAGCTCCATGACTTGATCCTCCTCTCTC |
| CUST_17700_PI426222305 | JHI_St_60k_v1 | DMT400066803 | TGAGAAGGGAGGGTATCCTTTGAACAGTCTGTTACAACTTGCAAGAAATTATTCTGAAGT |
Microarray Signals from CUST_17712_PI426222305
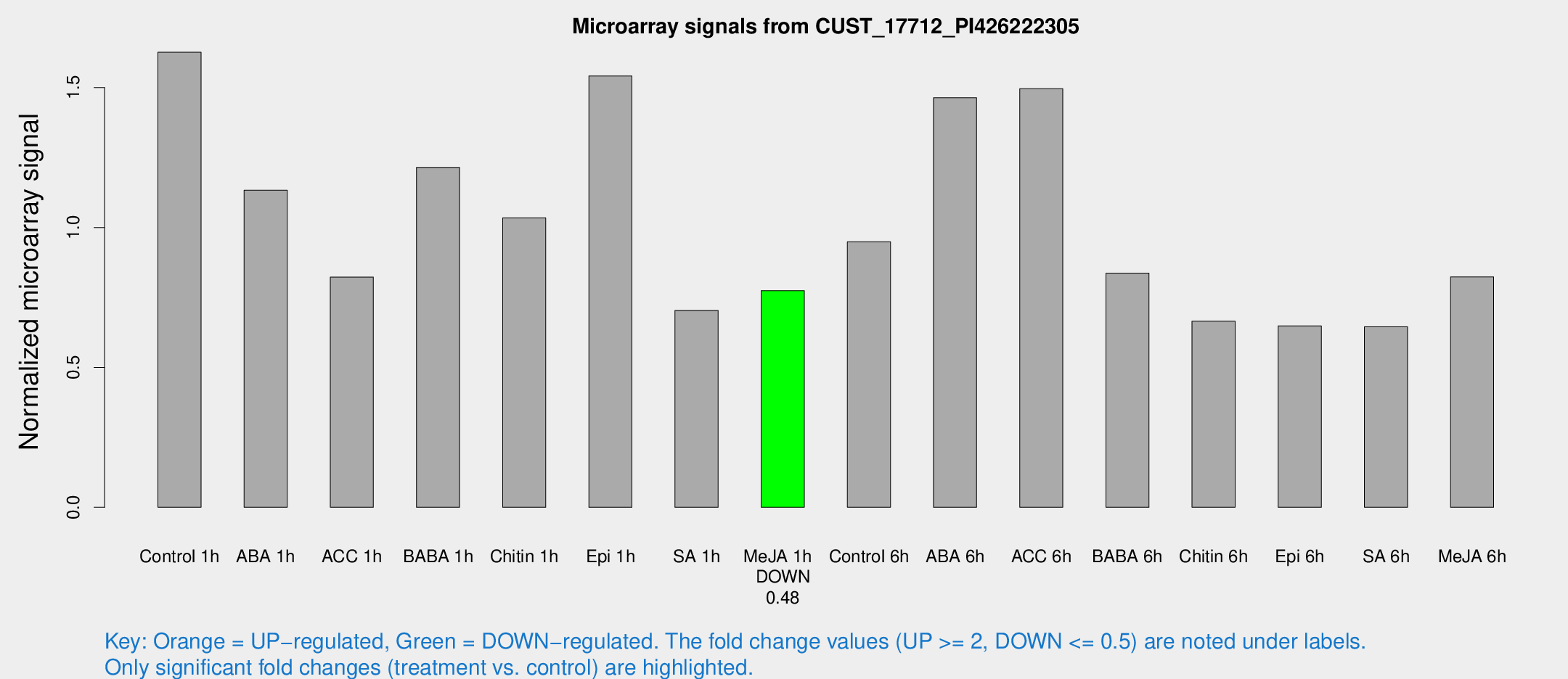
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.0302 | 3.13418 | 1.62649 | 0.270091 |
| ABA 1h | 12.8096 | 3.92229 | 1.13305 | 0.475339 |
| ACC 1h | 9.85534 | 3.27078 | 0.822573 | 0.275703 |
| BABA 1h | 14.2819 | 3.22992 | 1.21472 | 0.417572 |
| Chitin 1h | 11.8578 | 3.29917 | 1.03463 | 0.458746 |
| Epi 1h | 17.1672 | 5.62552 | 1.54156 | 0.522112 |
| SA 1h | 9.78688 | 3.98144 | 0.703749 | 0.350214 |
| Me-JA 1h | 7.57794 | 3.07902 | 0.774444 | 0.332014 |
| Control 6h | 12.3761 | 3.5291 | 0.949329 | 0.475737 |
| ABA 6h | 19.1852 | 4.83268 | 1.46356 | 0.387732 |
| ACC 6h | 23.5656 | 9.06025 | 1.49604 | 1.01144 |
| BABA 6h | 12.3165 | 4.44168 | 0.837042 | 0.303226 |
| Chitin 6h | 9.26525 | 3.45826 | 0.665307 | 0.313272 |
| Epi 6h | 9.56139 | 3.60181 | 0.648656 | 0.304785 |
| SA 6h | 8.12909 | 3.29731 | 0.645072 | 0.309789 |
| Me-JA 6h | 9.92405 | 3.17644 | 0.82363 | 0.286469 |
Source Transcript PGSC0003DMT400066804 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G30395.1 | -2 | 4e-31 | 119 | 62/93 (67%) | ovate family protein 17 | chr2:12954409-12955083 REVERSE LENGTH=195 |
