Probe CUST_17587_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17587_PI426222305 | JHI_St_60k_v1 | DMT400001527 | TATAGGTGAAAATTCGTCCAGTGAGATCAACATCGTCCGAGAGCCACTTCTAAAAGGCGA |
All Microarray Probes Designed to Gene DMG401000567
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17493_PI426222305 | JHI_St_60k_v1 | DMT400001526 | AATCTTCAATTTGACCTTCTCCAATGGCGACAGCCAACGATCCCGAGCAGCTAATTGTTT |
| CUST_17583_PI426222305 | JHI_St_60k_v1 | DMT400001525 | AATCTTCAATTTGACCTTCTCCAATGGCGACAGCCAACGATCCCGAGCAGCTAATTGTTT |
| CUST_17587_PI426222305 | JHI_St_60k_v1 | DMT400001527 | TATAGGTGAAAATTCGTCCAGTGAGATCAACATCGTCCGAGAGCCACTTCTAAAAGGCGA |
Microarray Signals from CUST_17587_PI426222305
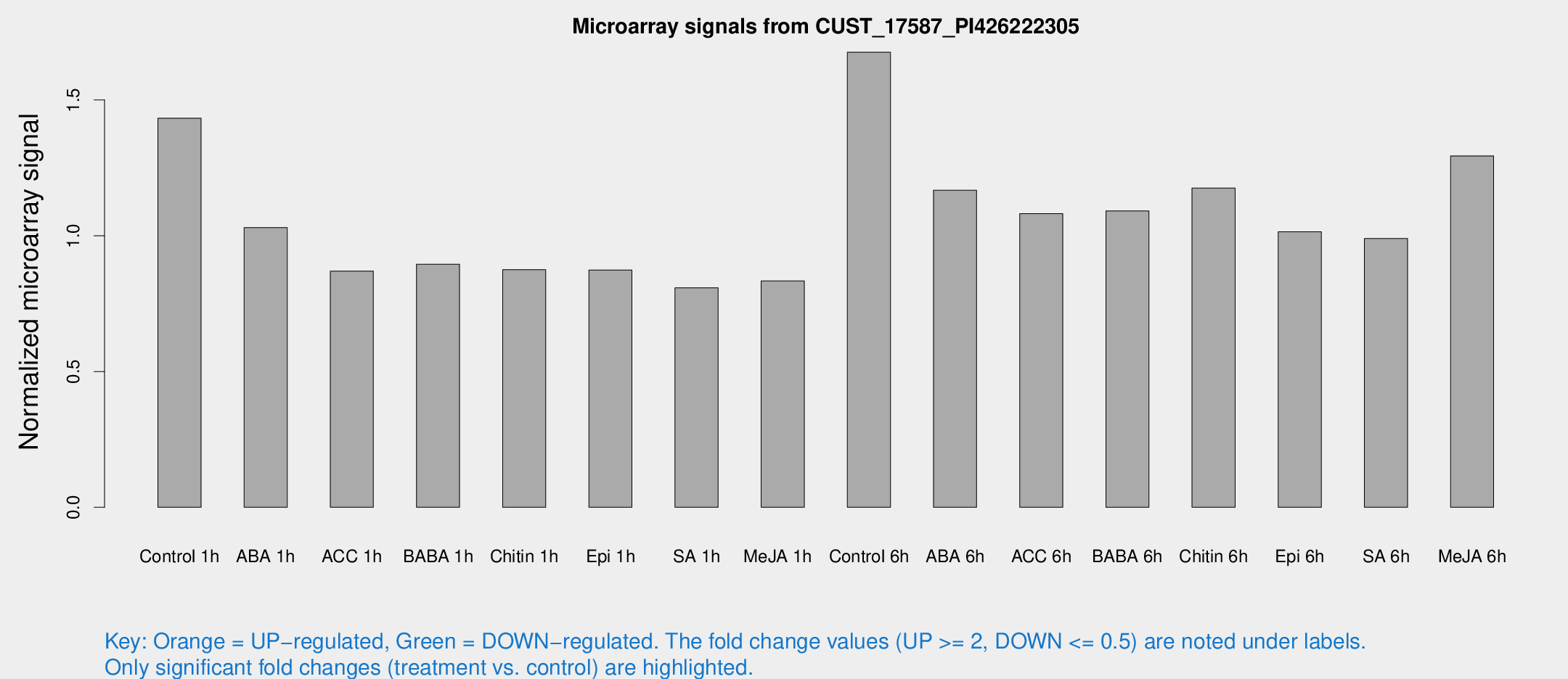
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 87.5644 | 6.01202 | 1.4328 | 0.0982349 |
| ABA 1h | 59.7788 | 17.1348 | 1.03008 | 0.31126 |
| ACC 1h | 59.2709 | 15.2559 | 0.869959 | 0.212197 |
| BABA 1h | 55.4606 | 13.5212 | 0.894974 | 0.191909 |
| Chitin 1h | 47.9661 | 4.388 | 0.875242 | 0.0799241 |
| Epi 1h | 47.0021 | 6.30838 | 0.873953 | 0.115137 |
| SA 1h | 54.0179 | 14.1692 | 0.808662 | 0.20081 |
| Me-JA 1h | 41.5802 | 4.17742 | 0.833885 | 0.0833983 |
| Control 6h | 114.714 | 33.2552 | 1.67588 | 0.482847 |
| ABA 6h | 78.9873 | 15.38 | 1.16734 | 0.218324 |
| ACC 6h | 76.8734 | 9.98832 | 1.08156 | 0.105906 |
| BABA 6h | 78.1954 | 15.788 | 1.09164 | 0.220102 |
| Chitin 6h | 77.0062 | 7.48294 | 1.17594 | 0.122624 |
| Epi 6h | 71.232 | 10.2301 | 1.01474 | 0.109601 |
| SA 6h | 60.9835 | 8.64511 | 0.989699 | 0.241736 |
| Me-JA 6h | 83.5416 | 19.5157 | 1.29409 | 0.229748 |
Source Transcript PGSC0003DMT400001527 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G17010.1 | +2 | 0.0 | 614 | 317/430 (74%) | Major facilitator superfamily protein | chr5:5587851-5592332 REVERSE LENGTH=503 |
