Probe CUST_16634_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16634_PI426222305 | JHI_St_60k_v1 | DMT400069678 | GTTGTATTCTTGTATGCAAATCAAAATACTCTGGGCAGAGGATAGACAAACCATTATATC |
All Microarray Probes Designed to Gene DMG400027088
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16568_PI426222305 | JHI_St_60k_v1 | DMT400069679 | GAAACATCAGGTACTGAAATTATCTGAAGTCTACACAGTTCTCCACAATTTCTAATCTTG |
| CUST_16782_PI426222305 | JHI_St_60k_v1 | DMT400069676 | CAACTGTCCTGATGGTATCTTCTTAATCCTATGATTTATGCACTTGCAGGATCAAGAACC |
| CUST_16634_PI426222305 | JHI_St_60k_v1 | DMT400069678 | GTTGTATTCTTGTATGCAAATCAAAATACTCTGGGCAGAGGATAGACAAACCATTATATC |
| CUST_16480_PI426222305 | JHI_St_60k_v1 | DMT400069675 | GTTGTATTCTTGTATGCAAATCAAAATACTCTGGGCAGAGGATAGACAAACCATTATATC |
| CUST_16708_PI426222305 | JHI_St_60k_v1 | DMT400069674 | TTGAAATAAAGCAAACTCCCAGCCCTCAGCCATCTAAACTTGGTCAAAATGATCGCGGTC |
Microarray Signals from CUST_16634_PI426222305
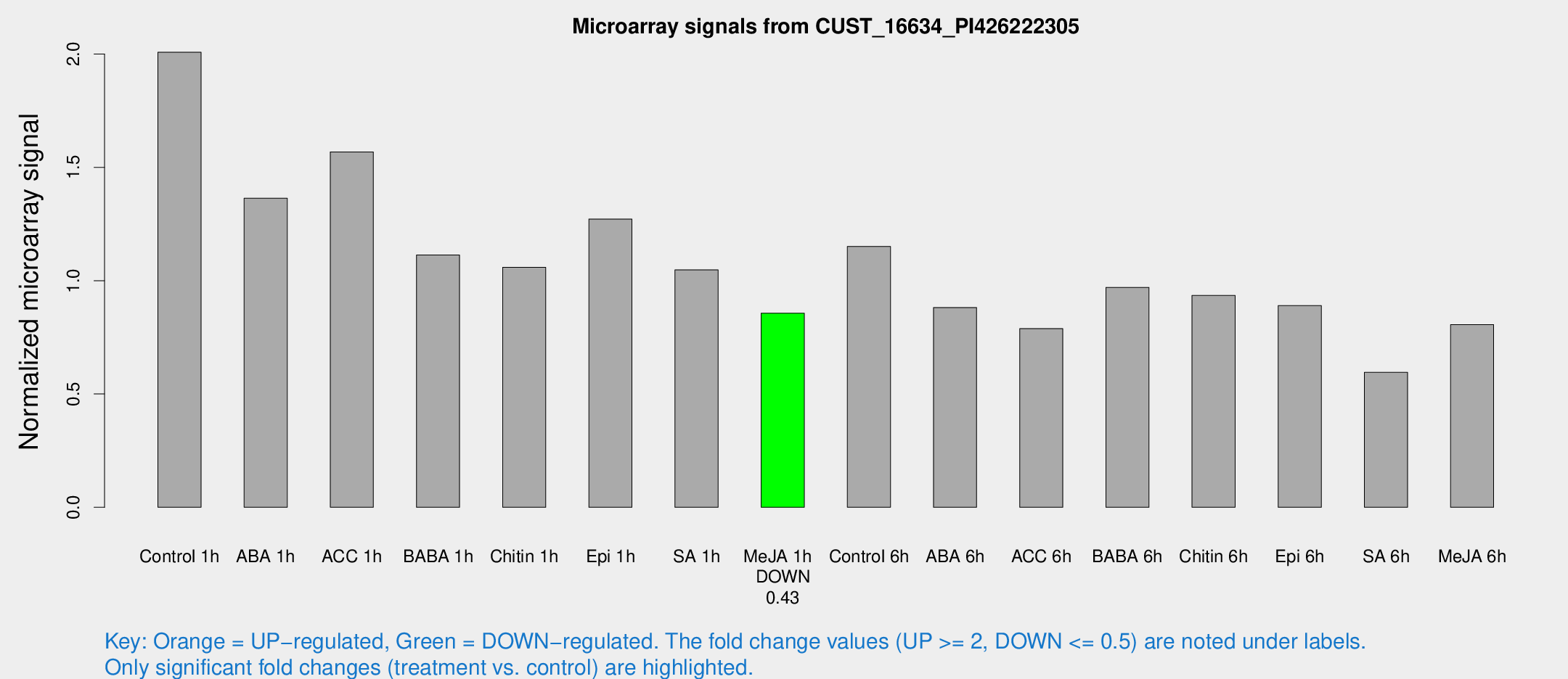
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3039.48 | 327.833 | 2.00798 | 0.115948 |
| ABA 1h | 1806.68 | 104.513 | 1.36403 | 0.145136 |
| ACC 1h | 2430.68 | 229.155 | 1.56827 | 0.0905711 |
| BABA 1h | 1639.5 | 233.319 | 1.1132 | 0.06431 |
| Chitin 1h | 1435.8 | 130.101 | 1.05902 | 0.0801675 |
| Epi 1h | 1680.13 | 234.727 | 1.27181 | 0.180026 |
| SA 1h | 1617.66 | 121.449 | 1.04764 | 0.0991494 |
| Me-JA 1h | 1047.97 | 60.739 | 0.85646 | 0.085847 |
| Control 6h | 1790.44 | 320.738 | 1.15098 | 0.128883 |
| ABA 6h | 1401.8 | 81.0311 | 0.881572 | 0.0582005 |
| ACC 6h | 1381.62 | 209.05 | 0.788504 | 0.0455733 |
| BABA 6h | 1637.82 | 150.798 | 0.970436 | 0.107501 |
| Chitin 6h | 1503.81 | 156.636 | 0.934829 | 0.0896965 |
| Epi 6h | 1506.7 | 103.306 | 0.889779 | 0.0906997 |
| SA 6h | 912.87 | 166.969 | 0.595489 | 0.0540701 |
| Me-JA 6h | 1249.64 | 248.315 | 0.806128 | 0.095645 |
Source Transcript PGSC0003DMT400069678 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G62270.1 | +3 | 0.0 | 1059 | 535/707 (76%) | HCO3- transporter family | chr3:23042528-23045633 REVERSE LENGTH=703 |
