Probe CUST_15853_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15853_PI426222305 | JHI_St_60k_v1 | DMT400036020 | CTTGGGTATTGGCCTTAATTCTTCATCAACATCAGATAATAATTGGAGTCATACCTTGAT |
All Microarray Probes Designed to Gene DMG400013875
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15933_PI426222305 | JHI_St_60k_v1 | DMT400036021 | CTTGGGTATTGGCCTTAATTCTTCATCAACATCAGATAATAATTGGAGTCATACCTTGAT |
| CUST_15853_PI426222305 | JHI_St_60k_v1 | DMT400036020 | CTTGGGTATTGGCCTTAATTCTTCATCAACATCAGATAATAATTGGAGTCATACCTTGAT |
| CUST_15866_PI426222305 | JHI_St_60k_v1 | DMT400036022 | CTTGGGTATTGGCCTTAATTCTTCATCAACATCAGATAATAATTGGAGTCATACCTTGAT |
Microarray Signals from CUST_15853_PI426222305
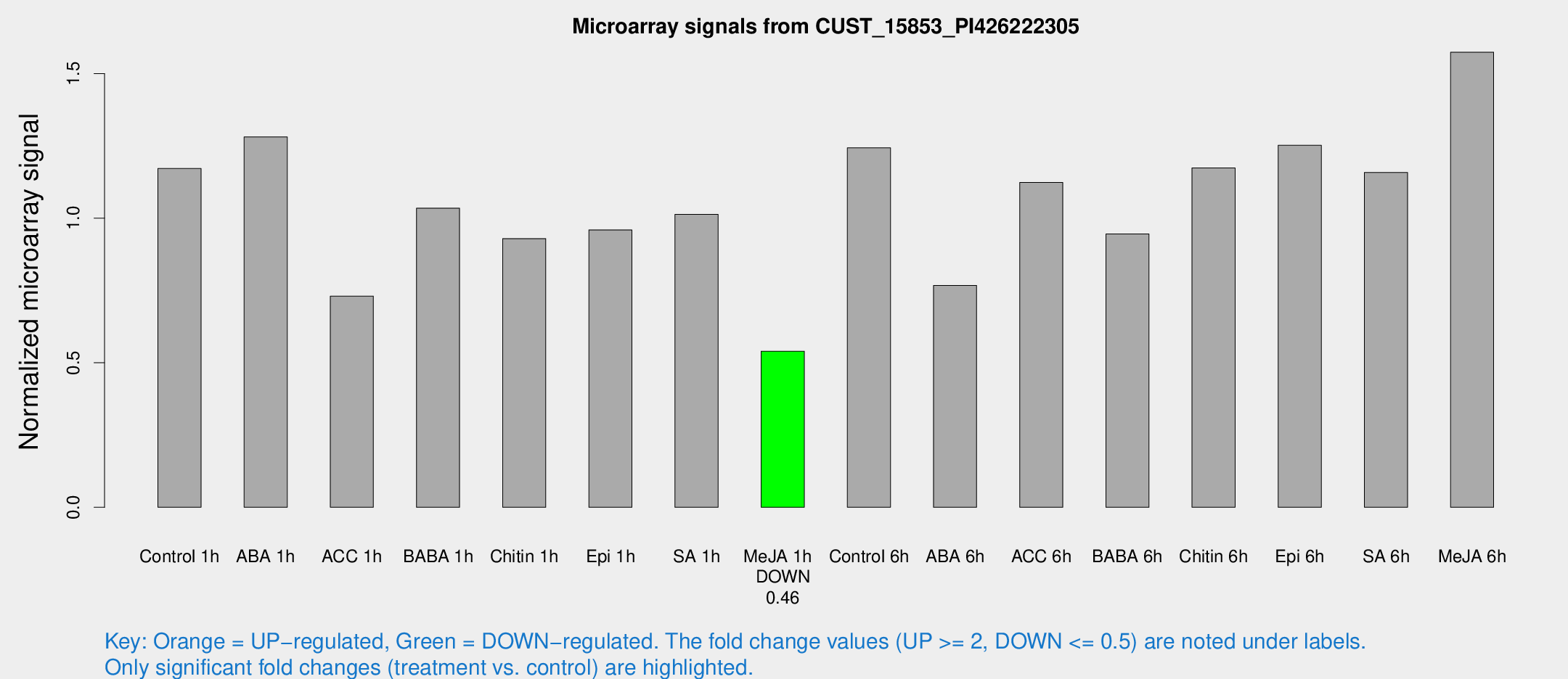
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 167.912 | 36.3874 | 1.17171 | 0.169146 |
| ABA 1h | 157.845 | 20.4008 | 1.28097 | 0.193034 |
| ACC 1h | 112.045 | 29.0469 | 0.730157 | 0.178409 |
| BABA 1h | 140.665 | 22.5928 | 1.03447 | 0.077795 |
| Chitin 1h | 119.53 | 25.5806 | 0.929325 | 0.122244 |
| Epi 1h | 113.853 | 7.36849 | 0.9592 | 0.0620759 |
| SA 1h | 145.779 | 20.3648 | 1.01304 | 0.0908415 |
| Me-JA 1h | 60.8005 | 4.92344 | 0.539879 | 0.0436586 |
| Control 6h | 192.626 | 60.5542 | 1.24325 | 0.370941 |
| ABA 6h | 113.411 | 12.9471 | 0.767267 | 0.0507863 |
| ACC 6h | 180.17 | 23.6428 | 1.12354 | 0.154897 |
| BABA 6h | 152.92 | 32.9293 | 0.94507 | 0.199966 |
| Chitin 6h | 180.459 | 40.6003 | 1.1735 | 0.302268 |
| Epi 6h | 195.56 | 19.6298 | 1.25206 | 0.186708 |
| SA 6h | 160.096 | 22.2499 | 1.1577 | 0.215759 |
| Me-JA 6h | 235.384 | 71.9223 | 1.5738 | 0.354407 |
Source Transcript PGSC0003DMT400036020 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G61660.1 | +1 | 2e-58 | 204 | 171/485 (35%) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | chr1:22754003-22756171 REVERSE LENGTH=393 |
