Probe CUST_15392_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15392_PI426222305 | JHI_St_60k_v1 | DMT400073685 | TGTTCTTTGACGATGTCCTACTCACAATGCATGATACATTTTAGATCTCTTCTTCCCTTG |
All Microarray Probes Designed to Gene DMG400028620
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15422_PI426222305 | JHI_St_60k_v1 | DMT400073683 | ACGATGTCCTACTCACAATGCATGATACATTTTAGATCTCTTCTTCCCTTGATTATGTTT |
| CUST_15392_PI426222305 | JHI_St_60k_v1 | DMT400073685 | TGTTCTTTGACGATGTCCTACTCACAATGCATGATACATTTTAGATCTCTTCTTCCCTTG |
| CUST_15458_PI426222305 | JHI_St_60k_v1 | DMT400073684 | ACGATGTCCTACTCACAATGCATGATACATTTTAGATCTCTTCTTCCCTTGATTATGTTT |
Microarray Signals from CUST_15392_PI426222305
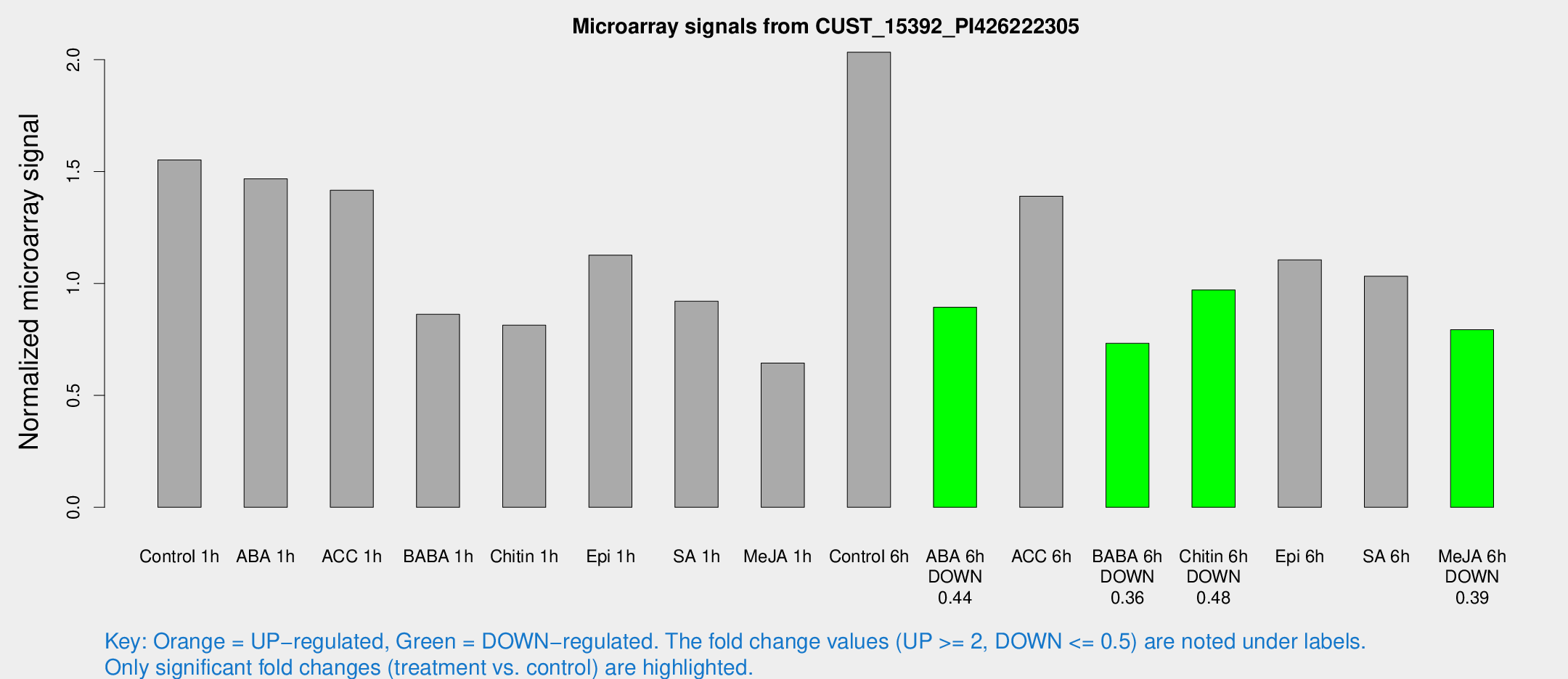
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 746.285 | 125.527 | 1.55215 | 0.160377 |
| ABA 1h | 652.058 | 169.393 | 1.4672 | 0.491831 |
| ACC 1h | 687.334 | 64.6165 | 1.41677 | 0.111124 |
| BABA 1h | 403.831 | 71.6203 | 0.862233 | 0.0807736 |
| Chitin 1h | 354.97 | 61.9085 | 0.813357 | 0.104935 |
| Epi 1h | 460.082 | 39.8624 | 1.12666 | 0.103604 |
| SA 1h | 455.197 | 78.3319 | 0.920486 | 0.158297 |
| Me-JA 1h | 247.761 | 17.9117 | 0.644439 | 0.0466862 |
| Control 6h | 983.989 | 175.177 | 2.03313 | 0.271269 |
| ABA 6h | 452.807 | 60.1777 | 0.893946 | 0.143744 |
| ACC 6h | 753.943 | 74.6668 | 1.39008 | 0.0805154 |
| BABA 6h | 387.065 | 34.5801 | 0.732494 | 0.0737883 |
| Chitin 6h | 486.79 | 36.1442 | 0.971124 | 0.0564689 |
| Epi 6h | 585.229 | 34.0392 | 1.10578 | 0.088666 |
| SA 6h | 502.142 | 115.722 | 1.03242 | 0.17958 |
| Me-JA 6h | 370.808 | 21.6659 | 0.7931 | 0.0462368 |
Source Transcript PGSC0003DMT400073685 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G15620.1 | +3 | 0.0 | 825 | 409/561 (73%) | DNA photolyase family protein | chr3:5293475-5296548 REVERSE LENGTH=556 |
