Probe CUST_15026_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15026_PI426222305 | JHI_St_60k_v1 | DMT400057318 | CTAAGTCTCTCTTTTCCAGTTACTCAGAGTTTTTAACTTTAGAGAGCAGTGTCTGATTAT |
All Microarray Probes Designed to Gene DMG400022258
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15002_PI426222305 | JHI_St_60k_v1 | DMT400057320 | CTAAGTCTCTCTTTTCCAGTTACTCAGAGTTTTTAACTTTAGAGAGCAGTGTCTGATTAT |
| CUST_15026_PI426222305 | JHI_St_60k_v1 | DMT400057318 | CTAAGTCTCTCTTTTCCAGTTACTCAGAGTTTTTAACTTTAGAGAGCAGTGTCTGATTAT |
| CUST_15292_PI426222305 | JHI_St_60k_v1 | DMT400057319 | ATTCTGCATTTTGCTGACCTGATTGTAACCAATGCAGGGGAATTGATCAATCTTCCTAAA |
Microarray Signals from CUST_15026_PI426222305
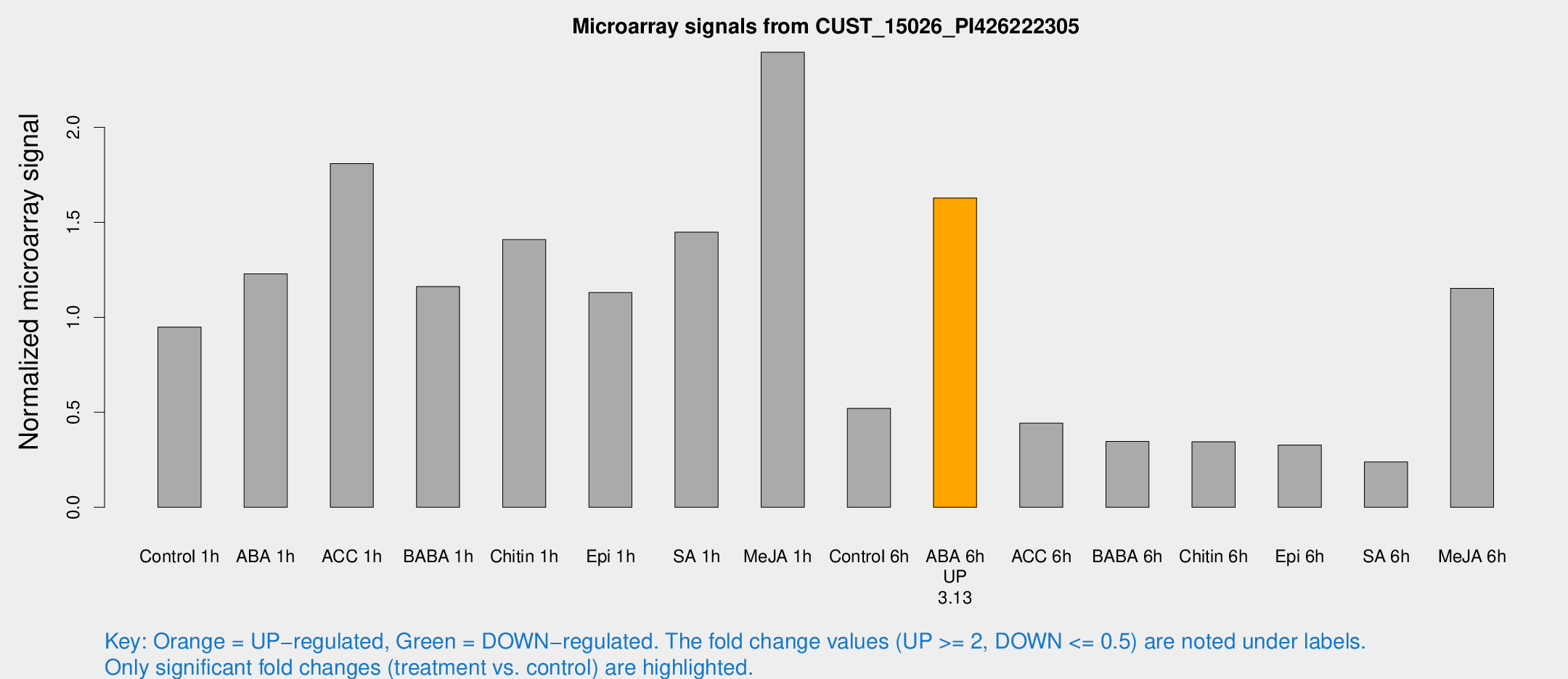
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 257.647 | 28.1288 | 0.94842 | 0.0652083 |
| ABA 1h | 300.957 | 54.6701 | 1.22859 | 0.148302 |
| ACC 1h | 541.544 | 146.557 | 1.80856 | 0.734567 |
| BABA 1h | 321.48 | 73.6991 | 1.16179 | 0.196921 |
| Chitin 1h | 373.383 | 119.986 | 1.40866 | 0.463909 |
| Epi 1h | 262.983 | 15.4865 | 1.13035 | 0.0665224 |
| SA 1h | 407.319 | 55.211 | 1.44812 | 0.163613 |
| Me-JA 1h | 548.233 | 104.675 | 2.39496 | 0.383673 |
| Control 6h | 158.716 | 58.077 | 0.520641 | 0.151408 |
| ABA 6h | 477.194 | 80.5269 | 1.62768 | 0.181049 |
| ACC 6h | 139.055 | 19.4806 | 0.442718 | 0.0327449 |
| BABA 6h | 105.055 | 9.25235 | 0.346788 | 0.0230655 |
| Chitin 6h | 99.1413 | 7.92925 | 0.344438 | 0.0374587 |
| Epi 6h | 103.801 | 20.6911 | 0.327228 | 0.0972955 |
| SA 6h | 74.2874 | 23.9614 | 0.238668 | 0.0816794 |
| Me-JA 6h | 313.238 | 38.3021 | 1.15273 | 0.251183 |
Source Transcript PGSC0003DMT400057318 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55740.1 | +3 | 2e-106 | 332 | 157/244 (64%) | seed imbibition 1 | chr1:20835507-20838707 REVERSE LENGTH=754 |
