Probe CUST_14751_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14751_PI426222305 | JHI_St_60k_v1 | DMT400066508 | CTCCGTCATAGGAAATTCTGATGCAACGGAATTGCGTTAAAAAACTGCTAATGATGAAGA |
All Microarray Probes Designed to Gene DMG400025844
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14751_PI426222305 | JHI_St_60k_v1 | DMT400066508 | CTCCGTCATAGGAAATTCTGATGCAACGGAATTGCGTTAAAAAACTGCTAATGATGAAGA |
Microarray Signals from CUST_14751_PI426222305
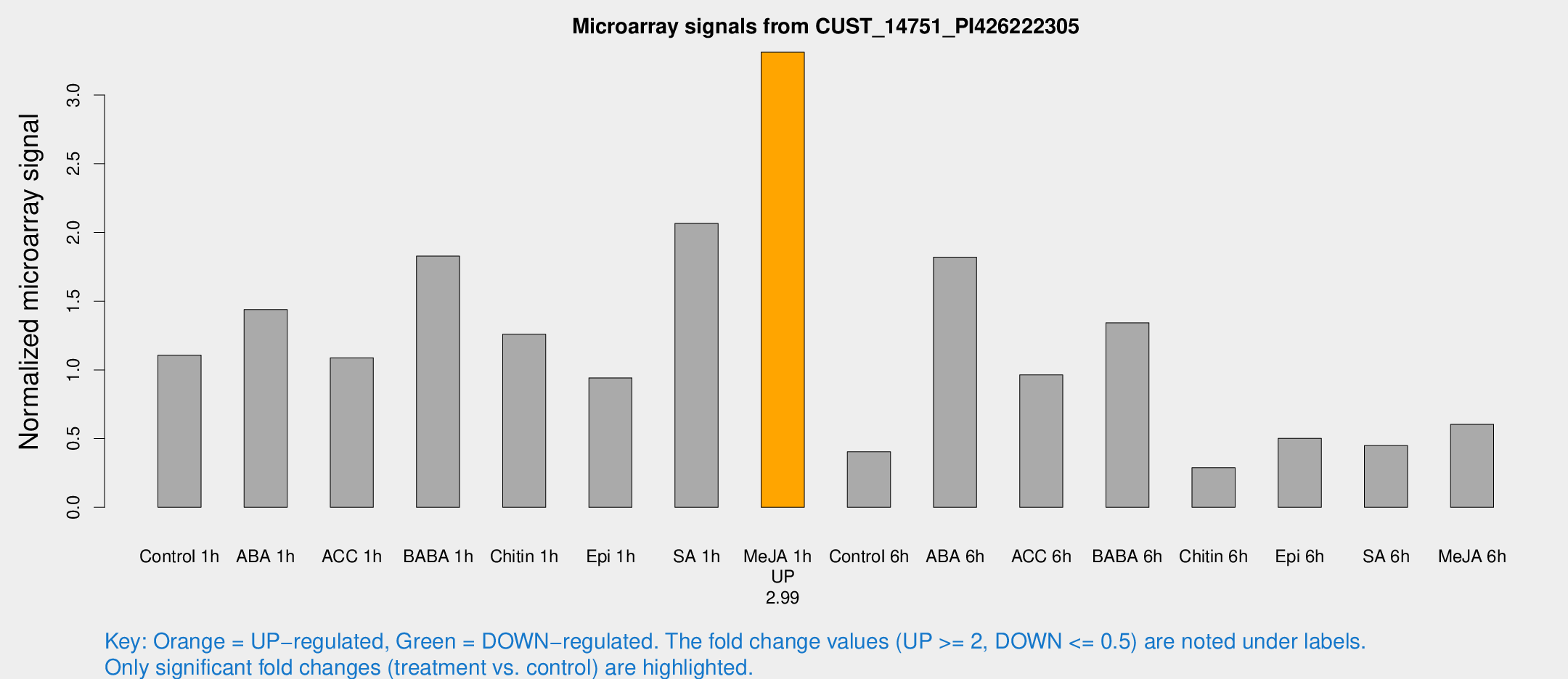
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.2168 | 6.93835 | 1.10782 | 0.139012 |
| ABA 1h | 43.0862 | 13.6558 | 1.43899 | 0.616525 |
| ACC 1h | 37.1785 | 9.59884 | 1.08823 | 0.256883 |
| BABA 1h | 59.0077 | 16.9894 | 1.82774 | 0.402854 |
| Chitin 1h | 35.5708 | 5.6668 | 1.25972 | 0.303612 |
| Epi 1h | 25.792 | 4.44985 | 0.942065 | 0.199488 |
| SA 1h | 70.2569 | 17.0801 | 2.06603 | 0.521701 |
| Me-JA 1h | 84.0155 | 9.65652 | 3.31188 | 0.227029 |
| Control 6h | 14.999 | 6.07519 | 0.404079 | 0.161017 |
| ABA 6h | 66.0691 | 21.5875 | 1.8199 | 0.526643 |
| ACC 6h | 34.258 | 4.25762 | 0.963731 | 0.225931 |
| BABA 6h | 50.5873 | 15.4603 | 1.34316 | 0.416647 |
| Chitin 6h | 9.70245 | 3.46875 | 0.288343 | 0.111846 |
| Epi 6h | 18.1163 | 3.83945 | 0.502361 | 0.107486 |
| SA 6h | 15.2352 | 4.87281 | 0.448926 | 0.187624 |
| Me-JA 6h | 19.3597 | 3.9448 | 0.603628 | 0.192306 |
Source Transcript PGSC0003DMT400066508 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G19615.1 | +2 | 3e-14 | 66 | 34/63 (54%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: cellular_component unknown; Has 10 Blast hits to 10 proteins in 5 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 10; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr3:6815073-6815354 REVERSE LENGTH=93 |
