Probe CUST_14509_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14509_PI426222305 | JHI_St_60k_v1 | DMT400066403 | TGGTCCAACTTTCAATGTAACTACAGTCTACACTAAGGTATTGCACTCGTTTTACTTGTC |
All Microarray Probes Designed to Gene DMG400025825
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14393_PI426222305 | JHI_St_60k_v1 | DMT400066402 | CTACAGTCTACACTAAGGATTTGAAGCTGGCTACTAAATTAGTAAGACCGCTATATTTAT |
| CUST_14581_PI426222305 | JHI_St_60k_v1 | DMT400066404 | GCCTTGTTGATGTTAGGATTTGAAGCTGGCTACTAAATTAGTAAGACCGCTATATTTATA |
| CUST_14662_PI426222305 | JHI_St_60k_v1 | DMT400066405 | CAACAACTTTTTACTACTCTTCTCCATATTGCCAATTCACATGCCTGATTTTAGTCCTAG |
| CUST_14509_PI426222305 | JHI_St_60k_v1 | DMT400066403 | TGGTCCAACTTTCAATGTAACTACAGTCTACACTAAGGTATTGCACTCGTTTTACTTGTC |
Microarray Signals from CUST_14509_PI426222305
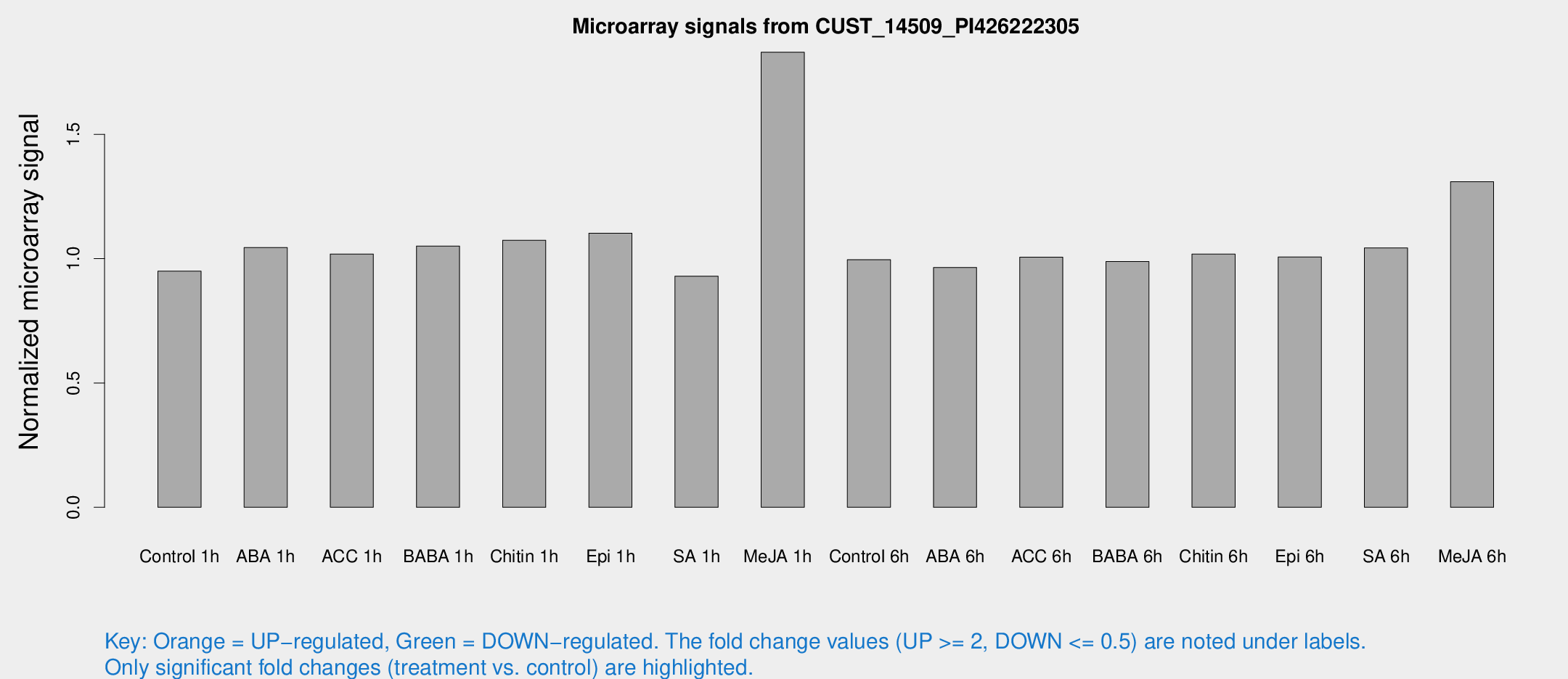
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.94875 | 3.44743 | 0.949838 | 0.550006 |
| ABA 1h | 5.79955 | 3.36778 | 1.04489 | 0.605206 |
| ACC 1h | 6.56348 | 3.81289 | 1.01865 | 0.590261 |
| BABA 1h | 6.33913 | 3.67142 | 1.05083 | 0.60856 |
| Chitin 1h | 6.0635 | 3.52211 | 1.07407 | 0.62198 |
| Epi 1h | 5.98301 | 3.47295 | 1.10251 | 0.638763 |
| SA 1h | 5.98051 | 3.46553 | 0.929674 | 0.53836 |
| Me-JA 1h | 11.269 | 5.08217 | 1.83012 | 0.875967 |
| Control 6h | 6.24406 | 3.61832 | 0.99547 | 0.576836 |
| ABA 6h | 6.42685 | 3.72684 | 0.964599 | 0.55856 |
| ACC 6h | 7.43629 | 4.43964 | 1.00602 | 0.582557 |
| BABA 6h | 6.9431 | 4.03212 | 0.988375 | 0.572398 |
| Chitin 6h | 6.78738 | 3.93242 | 1.01826 | 0.589599 |
| Epi 6h | 7.1518 | 4.16967 | 1.00688 | 0.583138 |
| SA 6h | 6.46104 | 3.74432 | 1.04298 | 0.603971 |
| Me-JA 6h | 8.39698 | 3.60034 | 1.30962 | 0.60157 |
Source Transcript PGSC0003DMT400066403 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23600.1 | +2 | 5e-37 | 138 | 65/129 (50%) | acetone-cyanohydrin lyase | chr2:10042325-10043641 REVERSE LENGTH=263 |
