Probe CUST_136_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_136_PI426222305 | JHI_St_60k_v1 | DMT400022528 | GTTTGGTTCGATTTCATGAATTTTGATGATGTAGAATAATTGGACGGATTGAGTTCTGAC |
All Microarray Probes Designed to Gene DMG400008734
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_136_PI426222305 | JHI_St_60k_v1 | DMT400022528 | GTTTGGTTCGATTTCATGAATTTTGATGATGTAGAATAATTGGACGGATTGAGTTCTGAC |
Microarray Signals from CUST_136_PI426222305
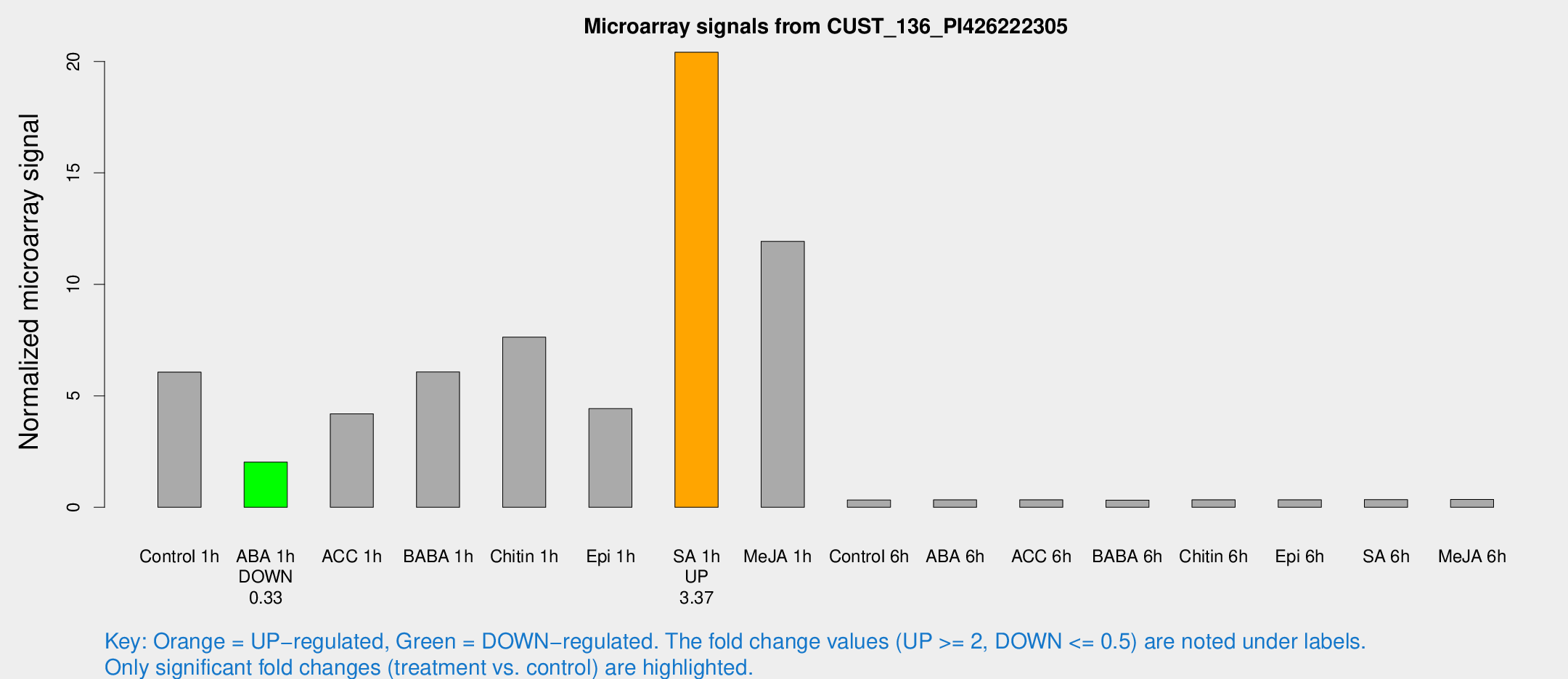
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 111.665 | 27.3161 | 6.06512 | 1.30671 |
| ABA 1h | 31.5268 | 3.78593 | 2.02549 | 0.378674 |
| ACC 1h | 79.2223 | 17.5301 | 4.19319 | 0.763521 |
| BABA 1h | 105.874 | 19.9174 | 6.07147 | 0.653018 |
| Chitin 1h | 119.169 | 7.59185 | 7.63158 | 0.78447 |
| Epi 1h | 66.6223 | 4.97826 | 4.4251 | 0.330219 |
| SA 1h | 367.363 | 36.3746 | 20.4128 | 1.24602 |
| Me-JA 1h | 174.548 | 32.5205 | 11.9317 | 1.31706 |
| Control 6h | 5.64017 | 3.26846 | 0.324204 | 0.187866 |
| ABA 6h | 6.20318 | 3.38813 | 0.334689 | 0.183781 |
| ACC 6h | 6.88342 | 4.09285 | 0.337108 | 0.195212 |
| BABA 6h | 6.30391 | 3.66275 | 0.323823 | 0.187797 |
| Chitin 6h | 6.26937 | 3.63147 | 0.339196 | 0.196403 |
| Epi 6h | 6.63982 | 3.8718 | 0.336963 | 0.195121 |
| SA 6h | 5.97854 | 3.46691 | 0.347966 | 0.201619 |
| Me-JA 6h | 6.06252 | 3.30262 | 0.348741 | 0.19136 |
Source Transcript PGSC0003DMT400022528 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc01g057080.1 | +2 | 5e-73 | 219 | 109/116 (94%) | evidence_code:10F0H1E0IEG genomic_reference:SL2.50ch01 gene_region:51195128-51195469 transcript_region:SL2.50ch01:51195128..51195469- go_terms:GO:0003677 functional_description:Ethylene-responsive transcription factor 13 (AHRD V1 *-*- ERF99_ARATH); contains Interpro domain(s) IPR001471 Pathogenesis-related transcriptional factor and ERF, DNA-binding |
| TAIR PP10 | AT1G71520.1 | +2 | 2e-37 | 129 | 64/126 (51%) | Integrase-type DNA-binding superfamily protein | chr1:26938612-26939043 FORWARD LENGTH=143 |
