Probe CUST_1127_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1127_PI426222305 | JHI_St_60k_v1 | DMT400001297 | AAGGCTTGGCAACTCTTCTTCTTCTTCTTCTATTCCAAGTCTCATCAGAAACGAGCCCGT |
All Microarray Probes Designed to Gene DMG400000493
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1127_PI426222305 | JHI_St_60k_v1 | DMT400001297 | AAGGCTTGGCAACTCTTCTTCTTCTTCTTCTATTCCAAGTCTCATCAGAAACGAGCCCGT |
Microarray Signals from CUST_1127_PI426222305
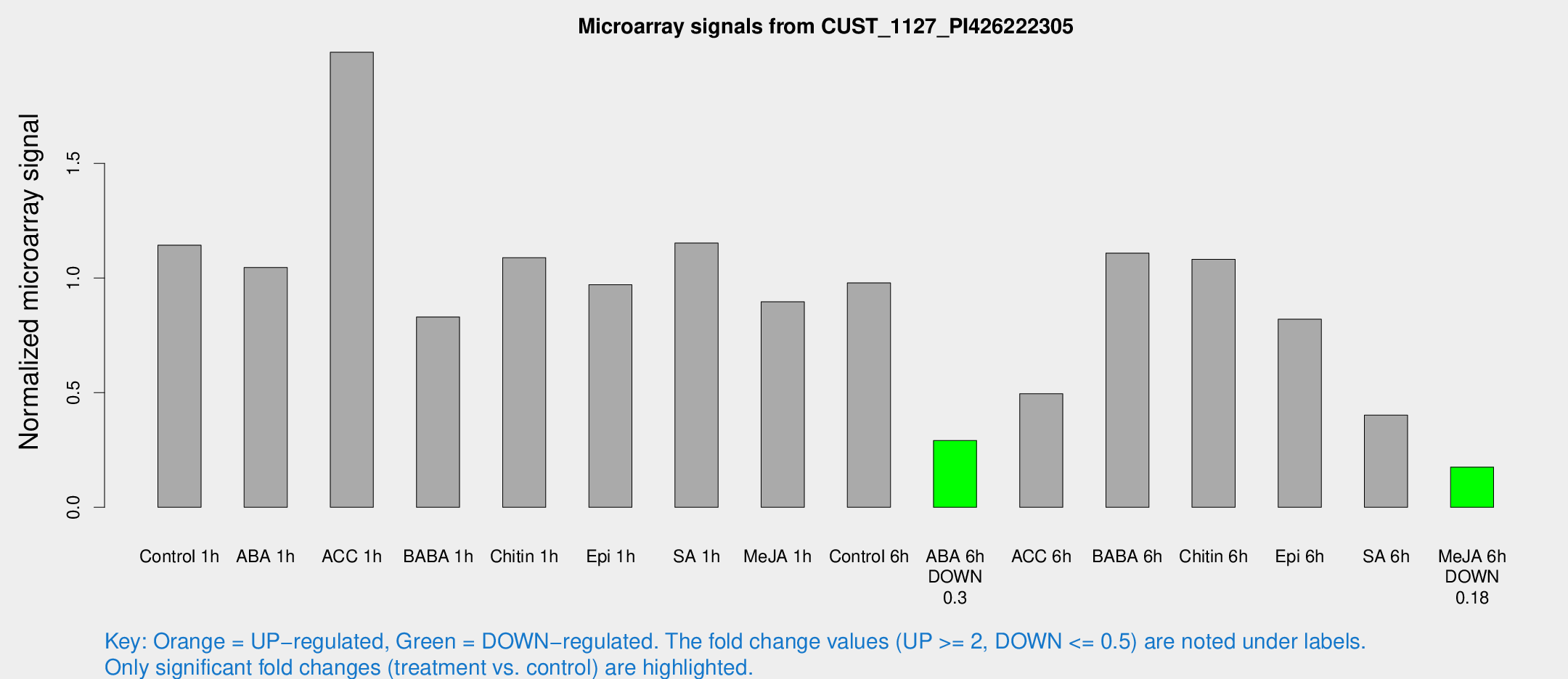
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7067.74 | 665.467 | 1.14325 | 0.0660085 |
| ABA 1h | 6253.65 | 1778.92 | 1.04603 | 0.404704 |
| ACC 1h | 13262.3 | 2974.87 | 1.98453 | 0.660561 |
| BABA 1h | 4912.3 | 284.223 | 0.829983 | 0.0617445 |
| Chitin 1h | 6558.69 | 2059.14 | 1.08868 | 0.360548 |
| Epi 1h | 5168.08 | 321.904 | 0.970738 | 0.0725453 |
| SA 1h | 7256.39 | 419.129 | 1.15231 | 0.0665309 |
| Me-JA 1h | 4758.52 | 1052.36 | 0.896047 | 0.320449 |
| Control 6h | 6866.32 | 2119.55 | 0.978755 | 0.311296 |
| ABA 6h | 1909.46 | 145.537 | 0.291326 | 0.0305935 |
| ACC 6h | 4178.43 | 1870.77 | 0.495171 | 0.144731 |
| BABA 6h | 8264.34 | 2206.74 | 1.10842 | 0.386628 |
| Chitin 6h | 7068.4 | 408.92 | 1.08155 | 0.0624465 |
| Epi 6h | 6153.4 | 1555.85 | 0.820365 | 0.340844 |
| SA 6h | 2759.75 | 1002.73 | 0.402301 | 0.147681 |
| Me-JA 6h | 1401.73 | 659.151 | 0.175341 | 0.0959774 |
Source Transcript PGSC0003DMT400001297 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g086820.2 | +2 | 0.0 | 561 | 317/322 (98%) | genomic_reference:SL2.50ch02 gene_region:44012475-44015926 transcript_region:SL2.50ch02:44012475..44015926+ go_terms:GO:0005515 functional_description:Carbonic anhydrase (AHRD V1 ***- Q5NE20_SOLLC); contains Interpro domain(s) IPR015892 Carbonic anhydrase, prokaryotic-like, conserved site |
| TAIR PP10 | AT3G01500.2 | +2 | 7e-131 | 389 | 221/318 (69%) | carbonic anhydrase 1 | chr3:194853-197873 REVERSE LENGTH=347 |
