Probe CUST_11266_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11266_PI426222305 | JHI_St_60k_v1 | DMT400037695 | CCTATTCAGATGGACAAAACAGAGGATTTGGAGACTAAAAAAGATGGGATTGAACTTGTA |
All Microarray Probes Designed to Gene DMG400014539
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11266_PI426222305 | JHI_St_60k_v1 | DMT400037695 | CCTATTCAGATGGACAAAACAGAGGATTTGGAGACTAAAAAAGATGGGATTGAACTTGTA |
| CUST_11069_PI426222305 | JHI_St_60k_v1 | DMT400037693 | CCTATTCAGATGGACAAAACAGAGGATTTGGAGACTAAAAAAGATGGGATTGAACTTGTA |
| CUST_11268_PI426222305 | JHI_St_60k_v1 | DMT400037694 | GGATCCTCTGAATATATTTTGGAGGATTGACATGAGTGTGTCAGCCTTTTTGAAGAGTAC |
Microarray Signals from CUST_11266_PI426222305
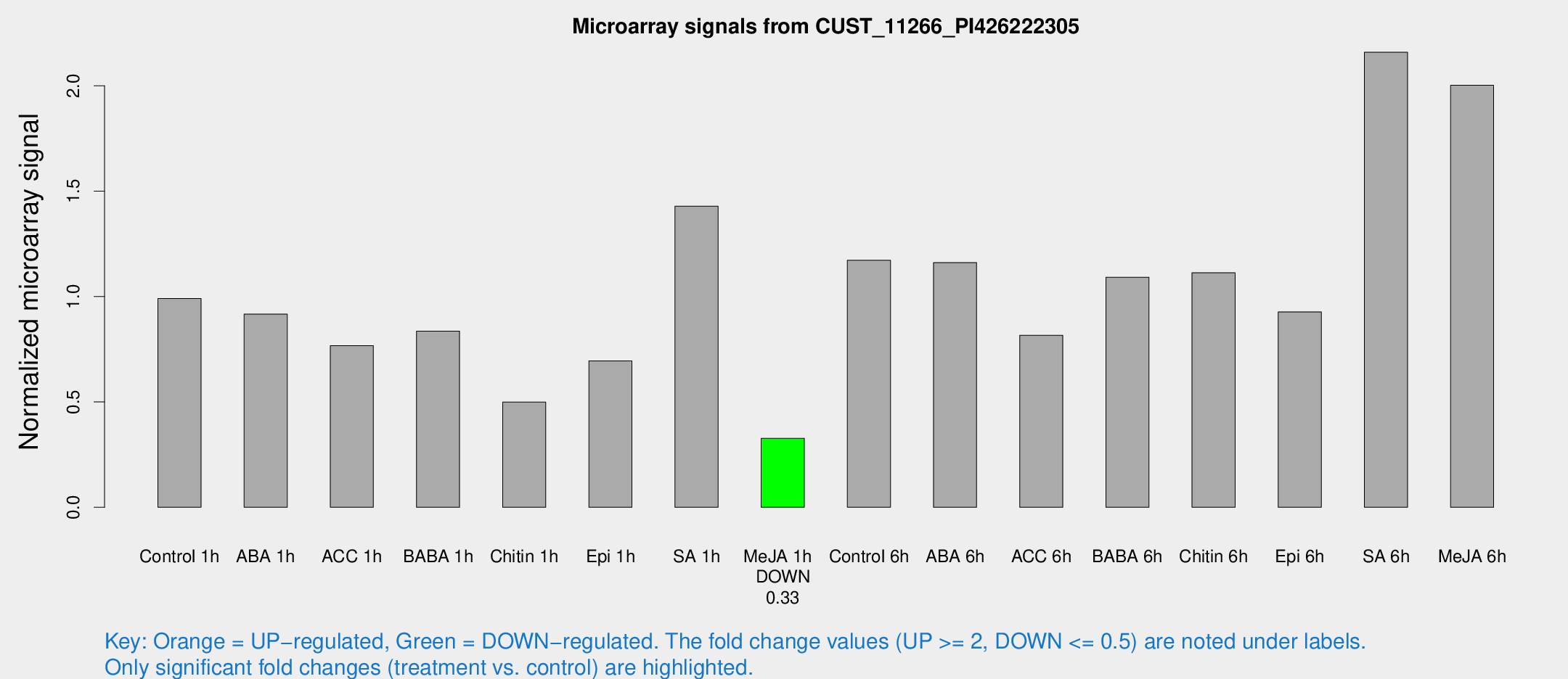
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5571.24 | 646.403 | 0.990508 | 0.0571893 |
| ABA 1h | 4716.21 | 924.2 | 0.916638 | 0.160131 |
| ACC 1h | 5280.86 | 1837.6 | 0.766655 | 0.351356 |
| BABA 1h | 4685.9 | 915.64 | 0.836119 | 0.0996595 |
| Chitin 1h | 2588.57 | 525.441 | 0.499321 | 0.0649245 |
| Epi 1h | 3495.91 | 686.086 | 0.695069 | 0.150882 |
| SA 1h | 8260.46 | 973.75 | 1.42906 | 0.0825084 |
| Me-JA 1h | 1518.19 | 233.291 | 0.327634 | 0.0311965 |
| Control 6h | 7205.46 | 2254.99 | 1.172 | 0.311641 |
| ABA 6h | 6859.36 | 396.335 | 1.16119 | 0.0670431 |
| ACC 6h | 5322.01 | 773.266 | 0.816537 | 0.0761564 |
| BABA 6h | 6898.6 | 888.521 | 1.09183 | 0.118529 |
| Chitin 6h | 6579.73 | 380.025 | 1.11292 | 0.0642567 |
| Epi 6h | 5864.45 | 605.126 | 0.927392 | 0.0535457 |
| SA 6h | 12838.9 | 3162.79 | 2.1595 | 0.39591 |
| Me-JA 6h | 11873.1 | 3255.28 | 2.00308 | 0.375477 |
Source Transcript PGSC0003DMT400037695 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68570.1 | +3 | 2e-148 | 439 | 214/294 (73%) | Major facilitator superfamily protein | chr1:25746811-25750110 FORWARD LENGTH=596 |
