Probe CUST_11013_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11013_PI426222305 | JHI_St_60k_v1 | DMT400037756 | ACCCGCACAAACTTCAACCCGCCCGCACAATTTTACTTTGTTAGATTGTAATTTGTAATA |
All Microarray Probes Designed to Gene DMG400014567
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11013_PI426222305 | JHI_St_60k_v1 | DMT400037756 | ACCCGCACAAACTTCAACCCGCCCGCACAATTTTACTTTGTTAGATTGTAATTTGTAATA |
Microarray Signals from CUST_11013_PI426222305
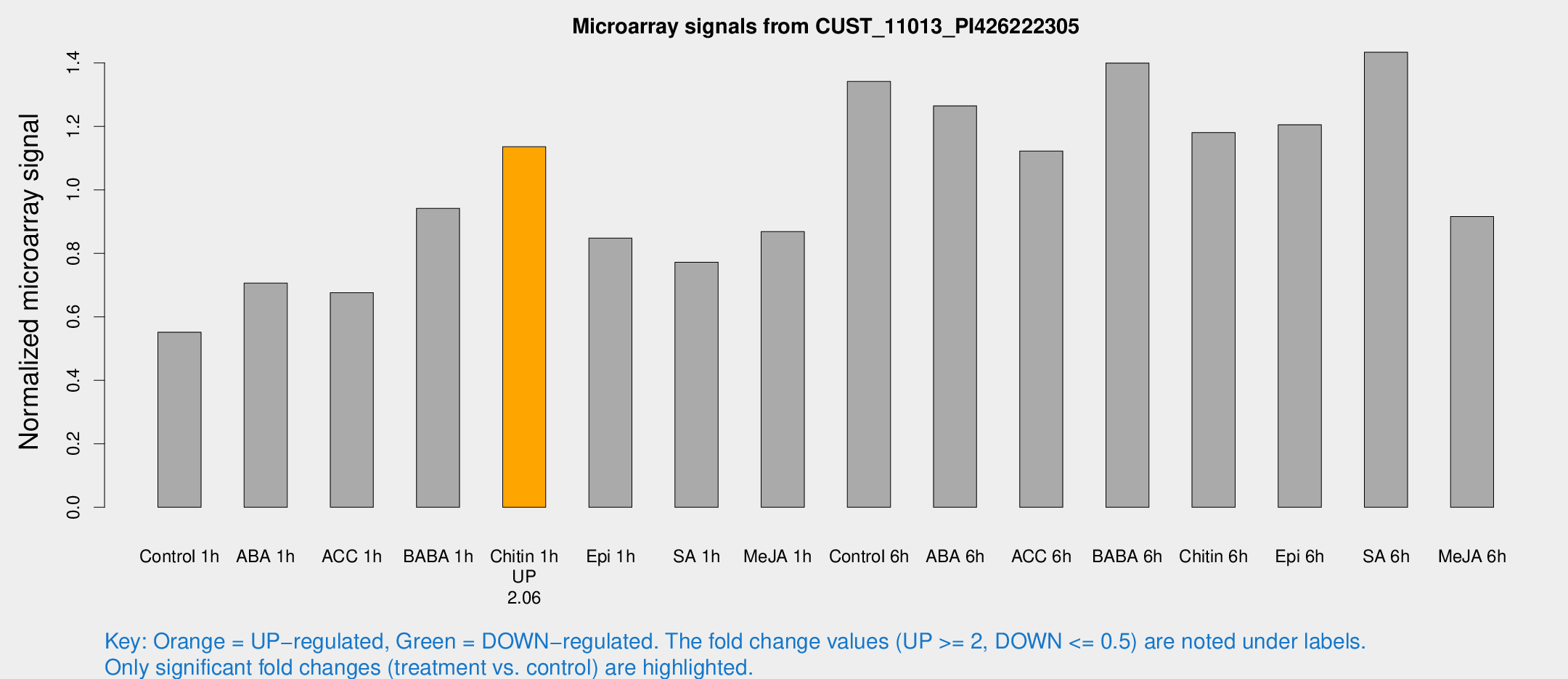
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2139.44 | 123.739 | 0.551625 | 0.0318571 |
| ABA 1h | 2494.03 | 450 | 0.706259 | 0.0876126 |
| ACC 1h | 2710.9 | 248.478 | 0.675853 | 0.0390291 |
| BABA 1h | 3629.24 | 601.054 | 0.941704 | 0.074038 |
| Chitin 1h | 3971.71 | 254.393 | 1.13583 | 0.0860742 |
| Epi 1h | 2882.57 | 340.308 | 0.848015 | 0.0803332 |
| SA 1h | 3139.78 | 480.259 | 0.772231 | 0.0729668 |
| Me-JA 1h | 2765.15 | 231.899 | 0.868707 | 0.0501637 |
| Control 6h | 5527.86 | 1209.52 | 1.34143 | 0.223676 |
| ABA 6h | 5245.3 | 440.042 | 1.26442 | 0.12281 |
| ACC 6h | 5036.78 | 514.235 | 1.12229 | 0.0648004 |
| BABA 6h | 6081.17 | 351.941 | 1.39944 | 0.0808005 |
| Chitin 6h | 4948.22 | 612.636 | 1.18045 | 0.132533 |
| Epi 6h | 5293.11 | 414.013 | 1.20506 | 0.118845 |
| SA 6h | 5494.62 | 317.436 | 1.43347 | 0.208908 |
| Me-JA 6h | 3837.09 | 970.533 | 0.916111 | 0.21285 |
Source Transcript PGSC0003DMT400037756 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G29180.1 | +3 | 5e-14 | 68 | 55/100 (55%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast thylakoid membrane; EXPRESSED IN: 23 plant structures; EXPRESSED DURING: 13 growth stages; Has 34 Blast hits to 33 proteins in 16 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 33; Viruses - 0; Other Eukaryotes - 1 (source: NCBI BLink). | chr2:12543081-12543702 FORWARD LENGTH=169 |
