Probe CUST_10417_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10417_PI426222305 | JHI_St_60k_v1 | DMT400029165 | TCCAAGAAATTTTCTTATTCAAGCTGTTGTTGGAGTTTTTGCTTTGGGGTTCATTGATGC |
All Microarray Probes Designed to Gene DMG400011214
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10417_PI426222305 | JHI_St_60k_v1 | DMT400029165 | TCCAAGAAATTTTCTTATTCAAGCTGTTGTTGGAGTTTTTGCTTTGGGGTTCATTGATGC |
Microarray Signals from CUST_10417_PI426222305
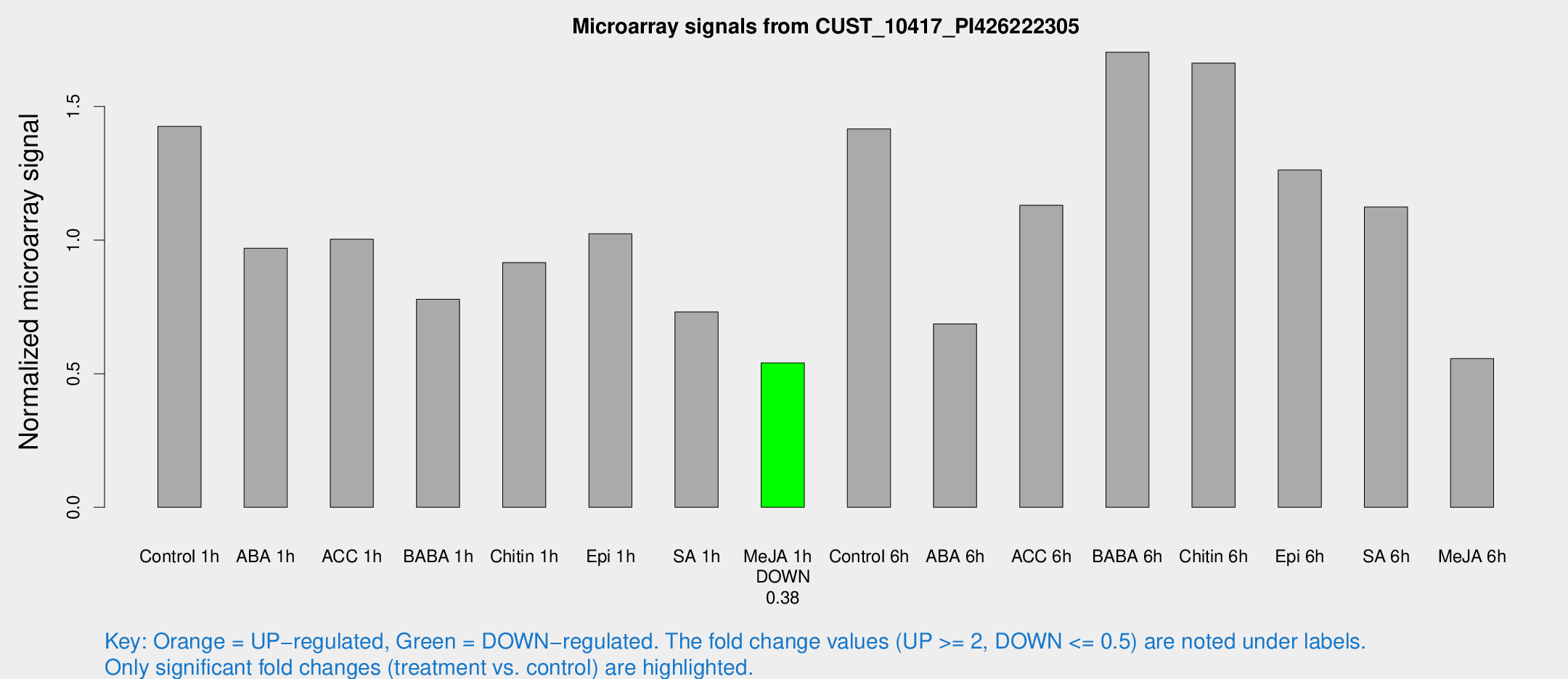
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 500.84 | 64.9856 | 1.42573 | 0.131226 |
| ABA 1h | 296.028 | 17.3772 | 0.969636 | 0.0568596 |
| ACC 1h | 359.522 | 39.5862 | 1.00347 | 0.0587979 |
| BABA 1h | 268.772 | 48.8442 | 0.77896 | 0.0782743 |
| Chitin 1h | 284.442 | 16.7575 | 0.91584 | 0.0538491 |
| Epi 1h | 307.959 | 27.667 | 1.02373 | 0.0747342 |
| SA 1h | 268.467 | 47.7624 | 0.731133 | 0.147181 |
| Me-JA 1h | 153.072 | 11.3577 | 0.540735 | 0.0332472 |
| Control 6h | 519.479 | 123.47 | 1.41607 | 0.258456 |
| ABA 6h | 253.021 | 18.4875 | 0.686452 | 0.065632 |
| ACC 6h | 474.546 | 109.394 | 1.1302 | 0.176125 |
| BABA 6h | 670.16 | 94.5359 | 1.70315 | 0.207255 |
| Chitin 6h | 630.655 | 119.209 | 1.66211 | 0.264075 |
| Epi 6h | 493.184 | 34.6796 | 1.26267 | 0.186672 |
| SA 6h | 401.254 | 78.563 | 1.12405 | 0.119243 |
| Me-JA 6h | 236.47 | 94.7366 | 0.556951 | 0.260064 |
Source Transcript PGSC0003DMT400029165 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G30845.1 | +2 | 6e-28 | 105 | 50/81 (62%) | unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: chloroplast; Has 16 Blast hits to 16 proteins in 8 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 16; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). | chr4:15019895-15020319 FORWARD LENGTH=114 |
