Probe CUST_8536_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8536_PI426222305 | JHI_St_60k_v1 | DMT400027660 | GAAACCGTAATATCGCTATAGTAATGAATAGGTAGTAGCTGAAAGTTGCAATCATTCAGT |
All Microarray Probes Designed to Gene DMG400010664
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8549_PI426222305 | JHI_St_60k_v1 | DMT400027659 | GAAACCGTAATATCGCTATAGTAATGAATAGGTAGTAGCTGAAAGTTGCAATCATTCAGT |
| CUST_8536_PI426222305 | JHI_St_60k_v1 | DMT400027660 | GAAACCGTAATATCGCTATAGTAATGAATAGGTAGTAGCTGAAAGTTGCAATCATTCAGT |
Microarray Signals from CUST_8536_PI426222305
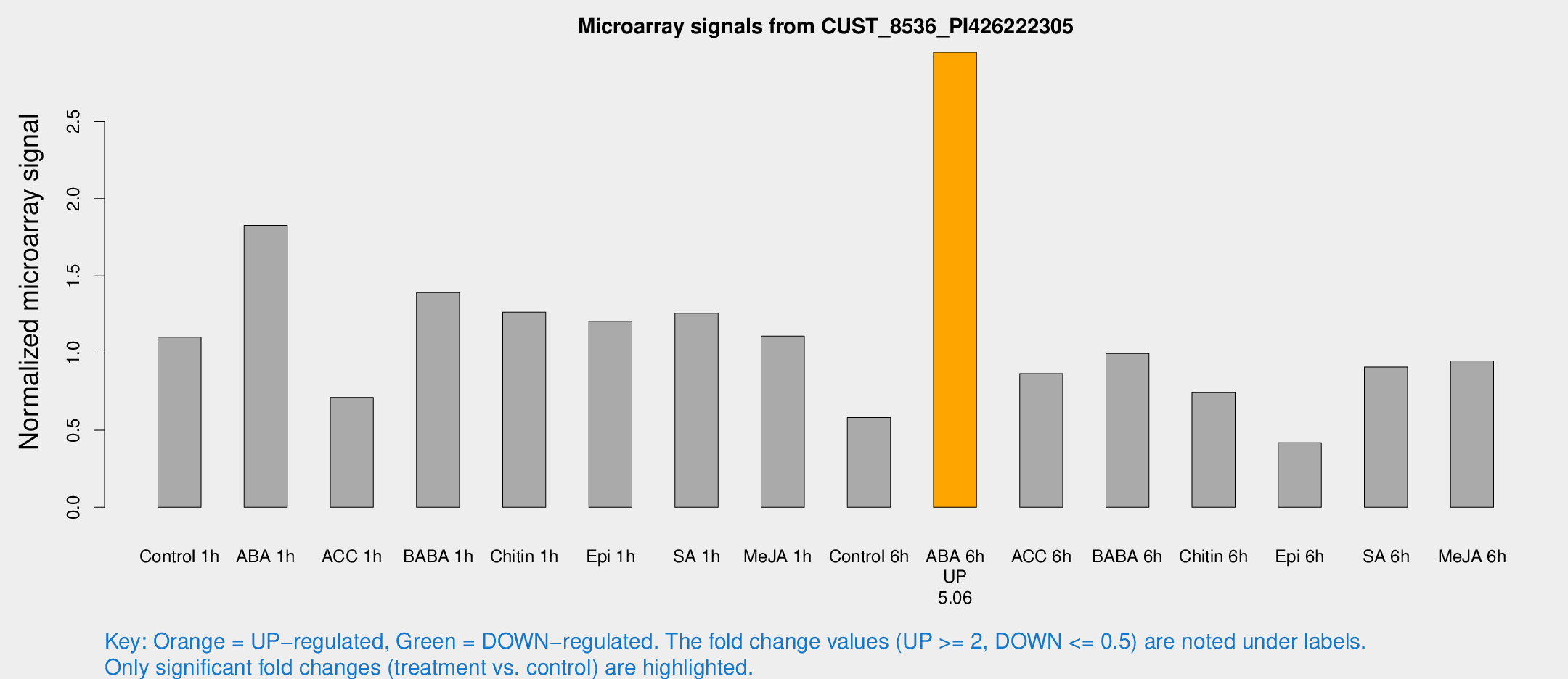
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1208.03 | 84.6532 | 1.10246 | 0.0637043 |
| ABA 1h | 1901.62 | 472.302 | 1.82778 | 0.468901 |
| ACC 1h | 924.015 | 314.449 | 0.712645 | 0.260428 |
| BABA 1h | 1560.55 | 354.448 | 1.39117 | 0.231461 |
| Chitin 1h | 1342.41 | 391.208 | 1.26451 | 0.300208 |
| Epi 1h | 1160.55 | 152.537 | 1.20645 | 0.157225 |
| SA 1h | 1508.63 | 411.087 | 1.2573 | 0.257099 |
| Me-JA 1h | 1021.41 | 194.332 | 1.10985 | 0.137618 |
| Control 6h | 681.948 | 178.263 | 0.582459 | 0.120899 |
| ABA 6h | 3447.31 | 322.161 | 2.94882 | 0.213242 |
| ACC 6h | 1091.31 | 95.5899 | 0.866199 | 0.0500861 |
| BABA 6h | 1253.92 | 216.752 | 0.997374 | 0.13503 |
| Chitin 6h | 863.096 | 50.005 | 0.742709 | 0.0429727 |
| Epi 6h | 526.157 | 83.3639 | 0.418187 | 0.0443816 |
| SA 6h | 1032.7 | 213.459 | 0.908981 | 0.109388 |
| Me-JA 6h | 1064.18 | 183.493 | 0.949262 | 0.0929719 |
Source Transcript PGSC0003DMT400027660 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18670.1 | +3 | 0.0 | 572 | 274/437 (63%) | beta-amylase 3 | chr5:6226138-6227999 FORWARD LENGTH=536 |
