Probe CUST_8202_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8202_PI426222305 | JHI_St_60k_v1 | DMT400075589 | GGTATCAAATGAAGTTGTAATCACTAAATTTTGTGGGGAGAAGATAGTTTTAGCTTGGTG |
All Microarray Probes Designed to Gene DMG400029396
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8351_PI426222305 | JHI_St_60k_v1 | DMT400075590 | GGTATCAAATGAAGTTGTAATCACTAAATTTTGTGGGGAGAAGATAGTTTTAGCTTGGTG |
| CUST_8202_PI426222305 | JHI_St_60k_v1 | DMT400075589 | GGTATCAAATGAAGTTGTAATCACTAAATTTTGTGGGGAGAAGATAGTTTTAGCTTGGTG |
Microarray Signals from CUST_8202_PI426222305
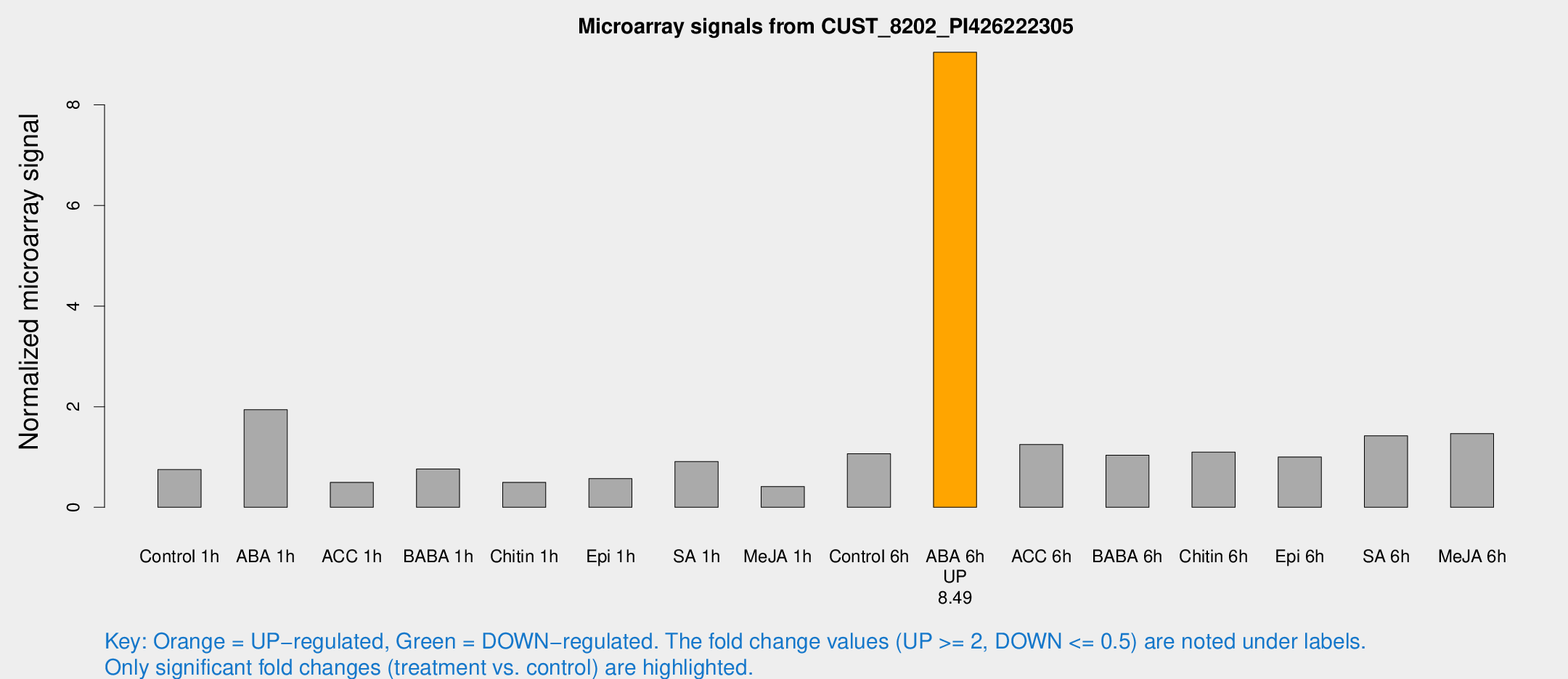
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 46.5809 | 8.76694 | 0.750806 | 0.122468 |
| ABA 1h | 109.185 | 29.1855 | 1.93969 | 0.503565 |
| ACC 1h | 34.145 | 10.9922 | 0.495333 | 0.15582 |
| BABA 1h | 46.1811 | 10.007 | 0.760233 | 0.113399 |
| Chitin 1h | 30.8759 | 10.4246 | 0.495863 | 0.228673 |
| Epi 1h | 32.3429 | 8.73021 | 0.571296 | 0.194811 |
| SA 1h | 56.5964 | 7.01743 | 0.910807 | 0.0758144 |
| Me-JA 1h | 20.8339 | 3.7028 | 0.413892 | 0.098832 |
| Control 6h | 66.0927 | 13.7933 | 1.06528 | 0.217787 |
| ABA 6h | 586.165 | 93.4287 | 9.04576 | 0.915551 |
| ACC 6h | 86.846 | 12.7215 | 1.24701 | 0.332931 |
| BABA 6h | 69.0514 | 5.21223 | 1.03556 | 0.0780343 |
| Chitin 6h | 69.4344 | 5.2051 | 1.09683 | 0.0822095 |
| Epi 6h | 74.1055 | 22.3939 | 1.00037 | 0.257744 |
| SA 6h | 92.2938 | 27.8424 | 1.42182 | 0.321138 |
| Me-JA 6h | 88.7779 | 13.6462 | 1.46638 | 0.117397 |
Source Transcript PGSC0003DMT400075589 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G52190.1 | +2 | 2e-139 | 426 | 235/466 (50%) | Major facilitator superfamily protein | chr1:19434671-19438673 FORWARD LENGTH=607 |
