Probe CUST_8094_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8094_PI426222305 | JHI_St_60k_v1 | DMT400030111 | TGCCACCATTCTCATTGAAATACAGGGCAGTAGTAAGGTCAAATTGTTTTCACAAGGAGG |
All Microarray Probes Designed to Gene DMG400011538
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8392_PI426222305 | JHI_St_60k_v1 | DMT400030112 | CCATTCTCATTGAAATACAGGCAAGAATTTTCGTATACGTATGTTGCACGTGTATATCAA |
| CUST_8094_PI426222305 | JHI_St_60k_v1 | DMT400030111 | TGCCACCATTCTCATTGAAATACAGGGCAGTAGTAAGGTCAAATTGTTTTCACAAGGAGG |
Microarray Signals from CUST_8094_PI426222305
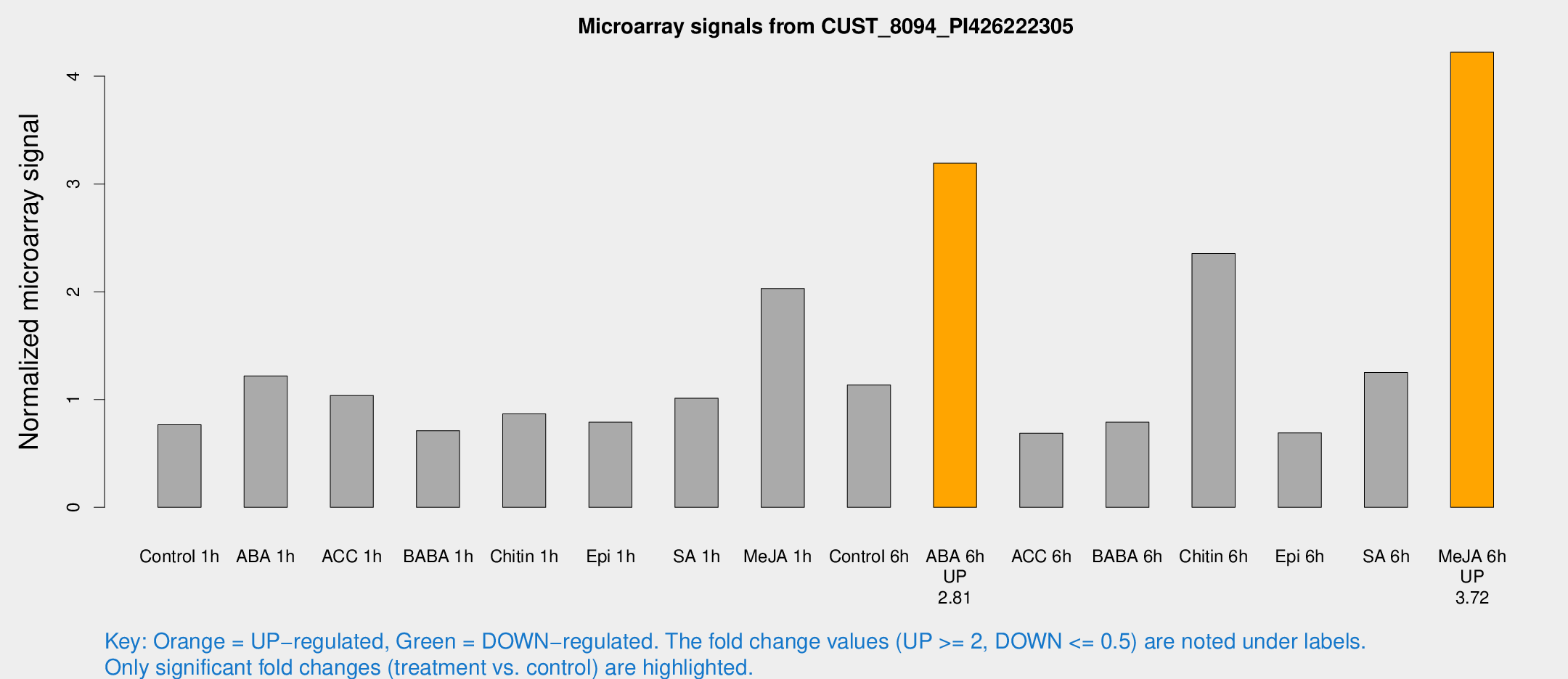
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.82298 | 3.72908 | 0.76631 | 0.385827 |
| ABA 1h | 11.2611 | 3.65131 | 1.21882 | 0.459411 |
| ACC 1h | 11.1558 | 4.04224 | 1.03684 | 0.419233 |
| BABA 1h | 6.83591 | 3.87595 | 0.711069 | 0.403816 |
| Chitin 1h | 7.79777 | 3.73893 | 0.866948 | 0.421886 |
| Epi 1h | 6.83185 | 3.61704 | 0.789482 | 0.422037 |
| SA 1h | 11.4754 | 3.86691 | 1.01216 | 0.404231 |
| Me-JA 1h | 18.0672 | 5.18056 | 2.02984 | 0.542109 |
| Control 6h | 11.7329 | 3.81124 | 1.13467 | 0.403162 |
| ABA 6h | 34.558 | 5.12539 | 3.19213 | 0.401046 |
| ACC 6h | 8.0352 | 4.74535 | 0.686303 | 0.393269 |
| BABA 6h | 9.04029 | 4.15237 | 0.789333 | 0.389279 |
| Chitin 6h | 36.465 | 22.5365 | 2.35458 | 1.7352 |
| Epi 6h | 7.81197 | 4.56254 | 0.690053 | 0.399595 |
| SA 6h | 13.5042 | 4.25061 | 1.25035 | 0.467045 |
| Me-JA 6h | 42.5426 | 5.44632 | 4.22274 | 0.45667 |
Source Transcript PGSC0003DMT400030111 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G49630.1 | +1 | 0.0 | 691 | 349/474 (74%) | amino acid permease 6 | chr5:20142681-20146441 REVERSE LENGTH=481 |
