Probe CUST_6999_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6999_PI426222305 | JHI_St_60k_v1 | DMT400031253 | GAGGTTAAAGATGGTGTCTTGTATATTACTATACCTAAGGCTAGTAGTAATCCCAAAGTT |
All Microarray Probes Designed to Gene DMG400011977
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6946_PI426222305 | JHI_St_60k_v1 | DMT400031252 | TGCAACAAATGATGGACACAATGGACAGAGTCATGGAGGACCCTCTTGCTTTTAATGGAG |
| CUST_6999_PI426222305 | JHI_St_60k_v1 | DMT400031253 | GAGGTTAAAGATGGTGTCTTGTATATTACTATACCTAAGGCTAGTAGTAATCCCAAAGTT |
Microarray Signals from CUST_6999_PI426222305
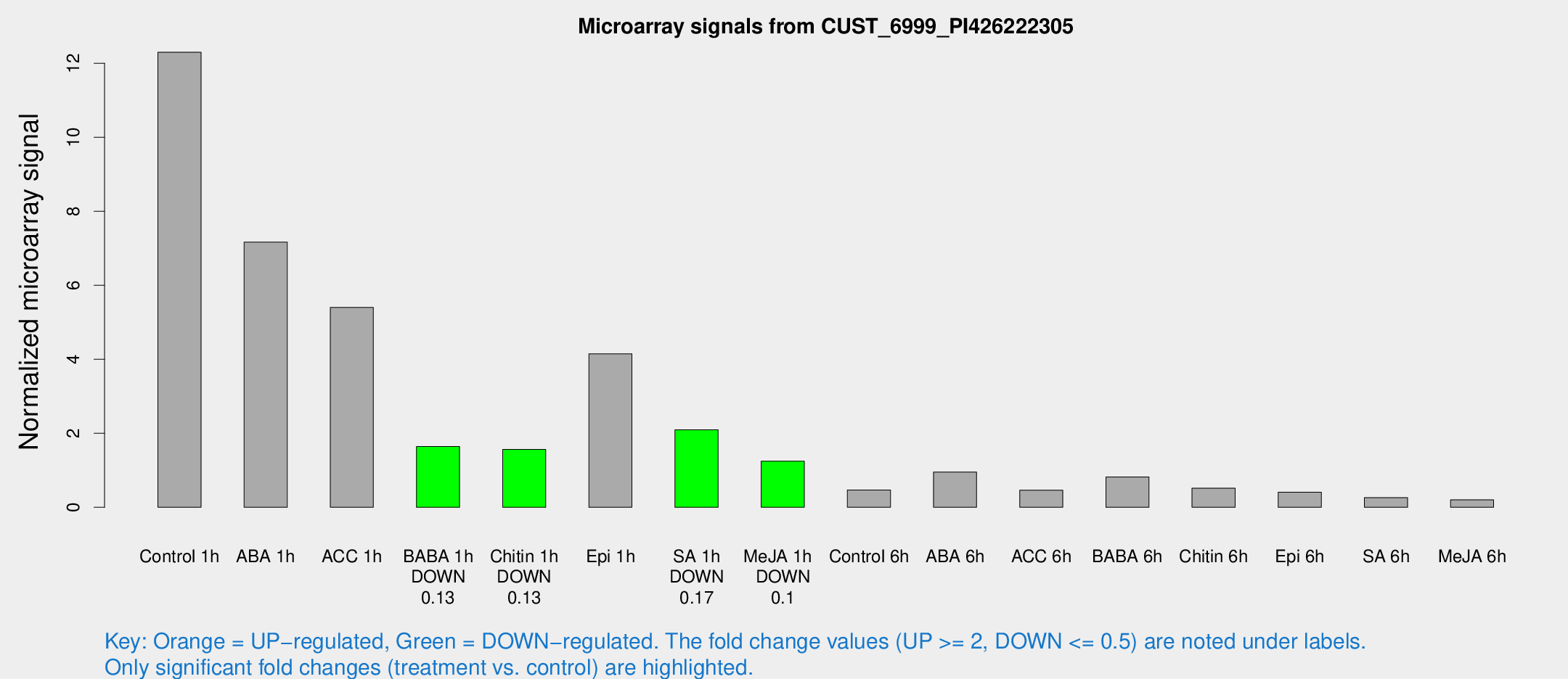
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 929.888 | 194.096 | 12.2976 | 2.42343 |
| ABA 1h | 773.597 | 539.656 | 7.16601 | 8.4694 |
| ACC 1h | 623.826 | 352.14 | 5.40208 | 5.19313 |
| BABA 1h | 134.833 | 52.7949 | 1.64321 | 0.589911 |
| Chitin 1h | 129.58 | 66.7376 | 1.56469 | 0.728169 |
| Epi 1h | 294.475 | 95.7213 | 4.14697 | 1.91491 |
| SA 1h | 161.83 | 31.4124 | 2.09507 | 0.371989 |
| Me-JA 1h | 73.3639 | 5.54086 | 1.24502 | 0.0941369 |
| Control 6h | 42.9755 | 16.018 | 0.469587 | 0.253617 |
| ABA 6h | 73.7645 | 7.38781 | 0.954281 | 0.122059 |
| ACC 6h | 38.312 | 4.9212 | 0.464217 | 0.0669548 |
| BABA 6h | 66.3634 | 5.55583 | 0.819062 | 0.0724913 |
| Chitin 6h | 40.0895 | 4.5652 | 0.517384 | 0.0599595 |
| Epi 6h | 34.4453 | 6.32966 | 0.40817 | 0.119622 |
| SA 6h | 18.8977 | 3.87658 | 0.260437 | 0.0572599 |
| Me-JA 6h | 17.161 | 6.63207 | 0.200092 | 0.0992517 |
Source Transcript PGSC0003DMT400031253 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27670.1 | +2 | 2e-33 | 119 | 55/119 (46%) | heat shock protein 21 | chr4:13819048-13819895 REVERSE LENGTH=227 |
