Probe CUST_6673_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6673_PI426222305 | JHI_St_60k_v1 | DMT400014383 | CTCAAGGAGAGAGTAGTACTCCTTTCCGAGACTTTTATTTGTTCAGTTTAGAAAATAAGA |
All Microarray Probes Designed to Gene DMG400005641
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_6673_PI426222305 | JHI_St_60k_v1 | DMT400014383 | CTCAAGGAGAGAGTAGTACTCCTTTCCGAGACTTTTATTTGTTCAGTTTAGAAAATAAGA |
| CUST_6395_PI426222305 | JHI_St_60k_v1 | DMT400014382 | TCATTCCAAATTGGGCAACAGGTATATTTCTTCTTTCAAATAAACCTGGCTGCTACCTCT |
Microarray Signals from CUST_6673_PI426222305
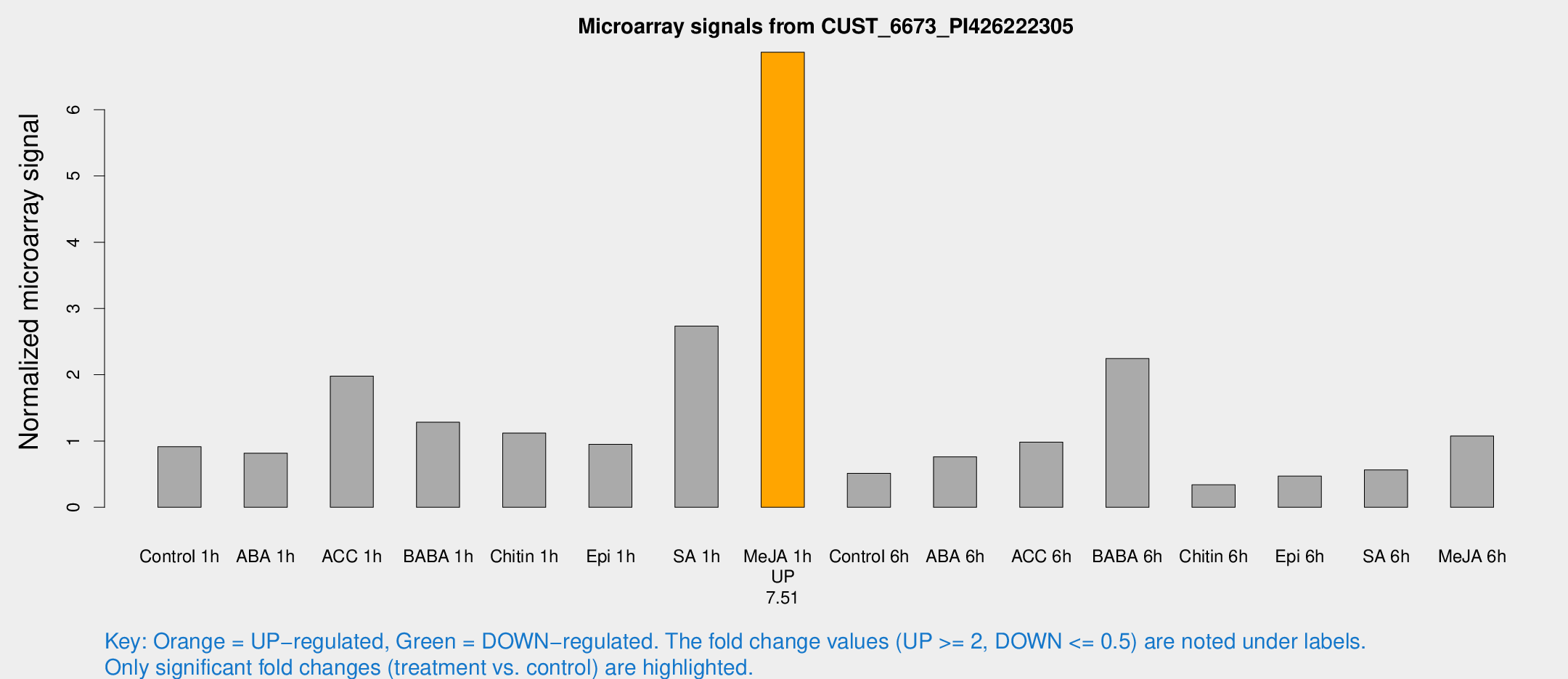
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 129.818 | 39.6903 | 0.913977 | 0.220583 |
| ABA 1h | 94.7964 | 6.21232 | 0.817672 | 0.0857994 |
| ACC 1h | 275.583 | 55.107 | 1.98034 | 0.405139 |
| BABA 1h | 164.321 | 20.9929 | 1.28427 | 0.0785044 |
| Chitin 1h | 140.092 | 34.7193 | 1.12014 | 0.365862 |
| Epi 1h | 108.208 | 9.51786 | 0.950274 | 0.113585 |
| SA 1h | 425.26 | 172.24 | 2.73507 | 1.13484 |
| Me-JA 1h | 749.863 | 114.423 | 6.86695 | 0.869185 |
| Control 6h | 73.2263 | 19.3682 | 0.512736 | 0.115218 |
| ABA 6h | 118.924 | 38.5648 | 0.762366 | 0.242482 |
| ACC 6h | 153.488 | 29.3988 | 0.984507 | 0.248848 |
| BABA 6h | 344.454 | 69.244 | 2.24534 | 0.566007 |
| Chitin 6h | 48.9333 | 9.06176 | 0.33921 | 0.0664897 |
| Epi 6h | 72.8382 | 15.0121 | 0.470406 | 0.0784619 |
| SA 6h | 73.6191 | 7.52509 | 0.564548 | 0.126232 |
| Me-JA 6h | 147.062 | 31.9058 | 1.07537 | 0.323636 |
Source Transcript PGSC0003DMT400014383 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68320.1 | +2 | 1e-72 | 230 | 106/131 (81%) | myb domain protein 62 | chr1:25603842-25604884 FORWARD LENGTH=286 |
