Probe CUST_5352_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5352_PI426222305 | JHI_St_60k_v1 | DMT400009094 | CAAGCAAAACTAACTGTGTTCTTCATTATTTACATCCTTAATCTCTCCAACATGTTGCTG |
All Microarray Probes Designed to Gene DMG400003536
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5396_PI426222305 | JHI_St_60k_v1 | DMT400009095 | CGTCGCTCTAGATCGCTTGCTGGTATGAGATCGCTTAACTTATATGGATTGACAGTATAA |
| CUST_5352_PI426222305 | JHI_St_60k_v1 | DMT400009094 | CAAGCAAAACTAACTGTGTTCTTCATTATTTACATCCTTAATCTCTCCAACATGTTGCTG |
Microarray Signals from CUST_5352_PI426222305
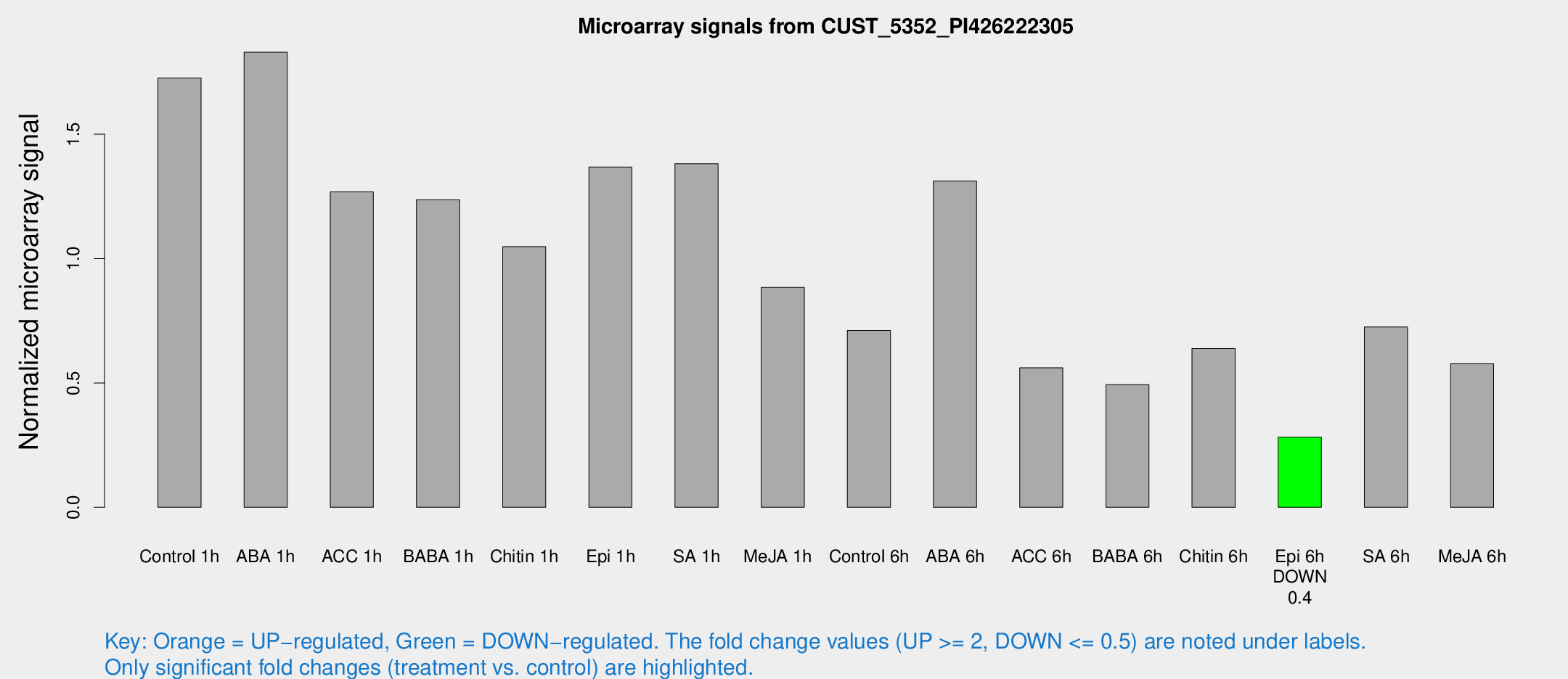
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 237.066 | 40.3458 | 1.72629 | 0.203528 |
| ABA 1h | 218.371 | 20.784 | 1.82994 | 0.159717 |
| ACC 1h | 191.313 | 52.8868 | 1.26835 | 0.323461 |
| BABA 1h | 165.283 | 29.4625 | 1.23609 | 0.117282 |
| Chitin 1h | 127.968 | 15.3692 | 1.04791 | 0.0682016 |
| Epi 1h | 162.355 | 24.2057 | 1.36825 | 0.204808 |
| SA 1h | 192.594 | 22.8032 | 1.38166 | 0.217584 |
| Me-JA 1h | 96.5934 | 6.58958 | 0.88439 | 0.0947588 |
| Control 6h | 100.008 | 22.9044 | 0.711377 | 0.13568 |
| ABA 6h | 187.088 | 11.5054 | 1.31211 | 0.0845442 |
| ACC 6h | 90.5153 | 18.8217 | 0.560832 | 0.122211 |
| BABA 6h | 73.9042 | 5.80077 | 0.493161 | 0.0388325 |
| Chitin 6h | 93.0671 | 15.108 | 0.638312 | 0.101769 |
| Epi 6h | 43.0308 | 5.25618 | 0.282422 | 0.0354608 |
| SA 6h | 97.3835 | 11.587 | 0.725408 | 0.144798 |
| Me-JA 6h | 77.648 | 8.1535 | 0.577019 | 0.0546749 |
Source Transcript PGSC0003DMT400009094 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36380.1 | +1 | 0.0 | 566 | 303/464 (65%) | Cytochrome P450 superfamily protein | chr4:17187973-17192202 REVERSE LENGTH=524 |
