Probe CUST_51625_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51625_PI426222305 | JHI_St_60k_v1 | DMT400053853 | AGAATGCTCTTTGGCGCGAAGAGGGAAGAAAGTTGAGGTTACTGTAAATTCCAAAATCTG |
All Microarray Probes Designed to Gene DMG400020895
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51625_PI426222305 | JHI_St_60k_v1 | DMT400053853 | AGAATGCTCTTTGGCGCGAAGAGGGAAGAAAGTTGAGGTTACTGTAAATTCCAAAATCTG |
| CUST_51620_PI426222305 | JHI_St_60k_v1 | DMT400053852 | TAAACTACAGAGCGAAGAGATTGCAAACACGATAAGGAAGGTGTTAGTTGAGGAAAGTGG |
Microarray Signals from CUST_51625_PI426222305
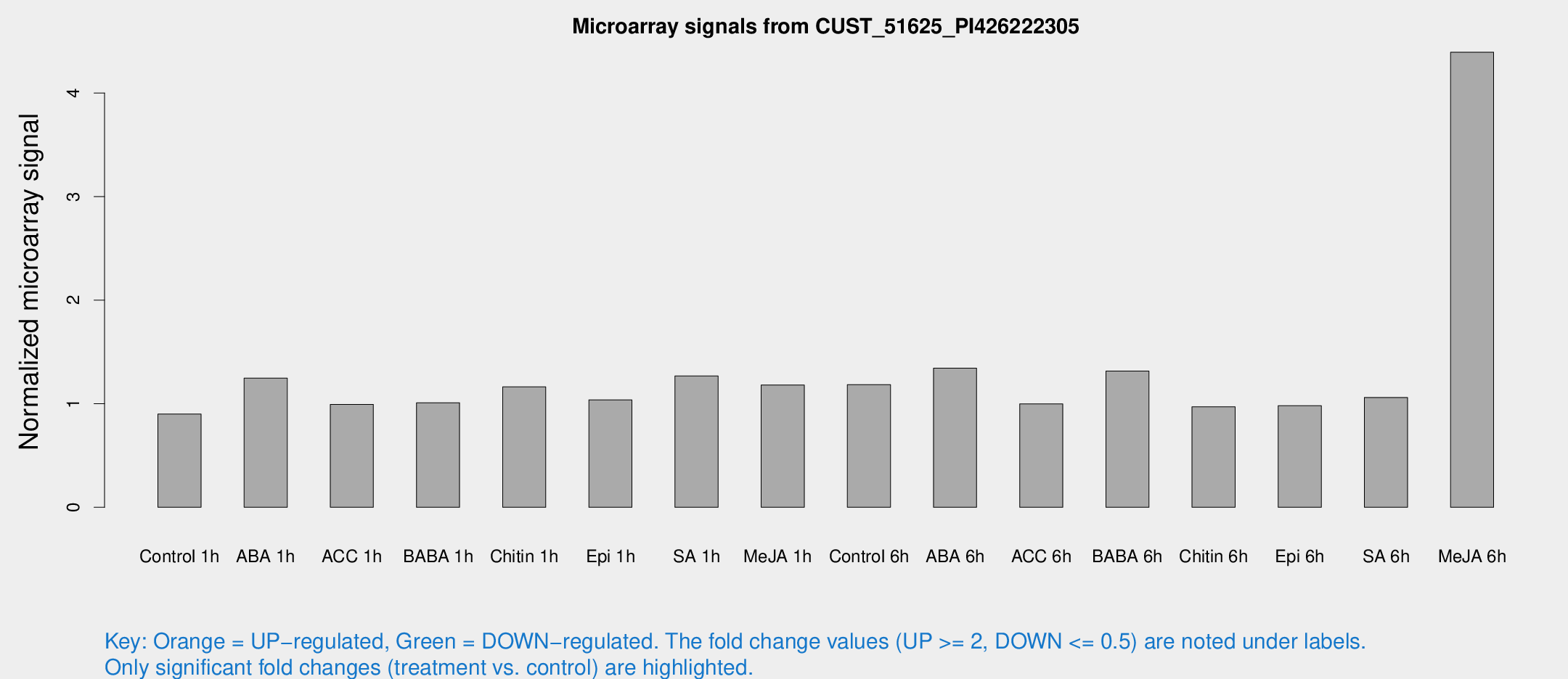
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.39307 | 3.70597 | 0.900383 | 0.521843 |
| ABA 1h | 8.24847 | 3.68528 | 1.24748 | 0.609301 |
| ACC 1h | 7.28118 | 4.24127 | 0.99306 | 0.575216 |
| BABA 1h | 6.90673 | 4.00124 | 1.00913 | 0.584535 |
| Chitin 1h | 7.60035 | 3.84434 | 1.1623 | 0.6073 |
| Epi 1h | 6.37833 | 3.69683 | 1.03712 | 0.60058 |
| SA 1h | 10.9968 | 4.79919 | 1.26784 | 0.591287 |
| Me-JA 1h | 6.8716 | 3.75985 | 1.18131 | 0.64656 |
| Control 6h | 8.82384 | 3.95878 | 1.18424 | 0.5842 |
| ABA 6h | 10.609 | 4.05756 | 1.34298 | 0.562866 |
| ACC 6h | 8.3069 | 4.78993 | 0.998229 | 0.559634 |
| BABA 6h | 12.4729 | 5.48878 | 1.31527 | 0.617822 |
| Chitin 6h | 7.33259 | 4.24719 | 0.969932 | 0.561614 |
| Epi 6h | 7.9068 | 4.61219 | 0.980363 | 0.567691 |
| SA 6h | 7.48496 | 4.08519 | 1.06057 | 0.583319 |
| Me-JA 6h | 55.9117 | 40.4112 | 4.39347 | 5.14226 |
Source Transcript PGSC0003DMT400053853 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65550.1 | +2 | 8e-48 | 128 | 68/158 (43%) | UDP-Glycosyltransferase superfamily protein | chr5:26198410-26199810 REVERSE LENGTH=466 |
