Probe CUST_51155_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51155_PI426222305 | JHI_St_60k_v1 | DMT400008555 | CTATCACAGTAAAACACTATTAGATCTGCTGATGTTGCAACTTGATTTTCGAAACTAAGC |
All Microarray Probes Designed to Gene DMG400003308
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51155_PI426222305 | JHI_St_60k_v1 | DMT400008555 | CTATCACAGTAAAACACTATTAGATCTGCTGATGTTGCAACTTGATTTTCGAAACTAAGC |
| CUST_51148_PI426222305 | JHI_St_60k_v1 | DMT400008556 | CTATCACAGTAAAACACTATTAGATCTGCTGATGTTGCAACTTGATTTTCGAAACTAAGC |
Microarray Signals from CUST_51155_PI426222305
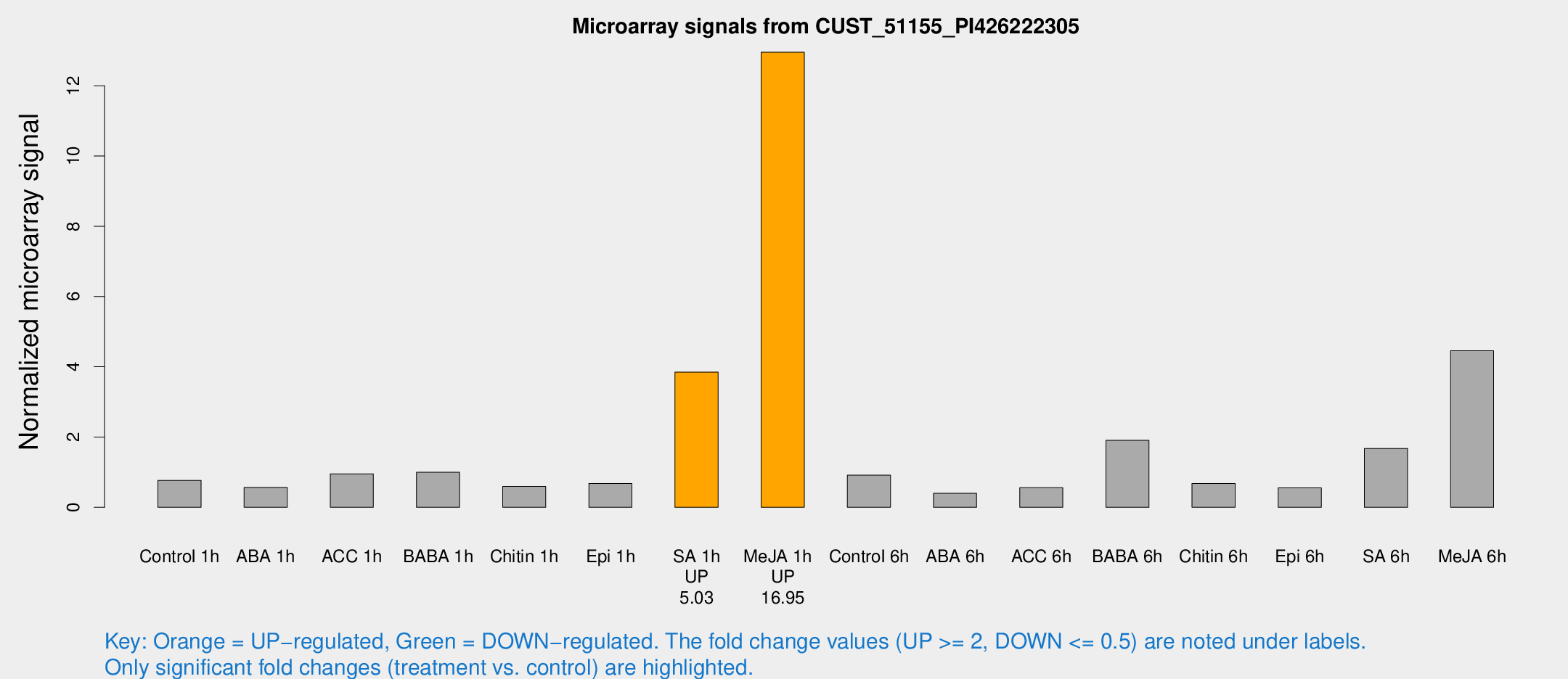
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.5801 | 8.97897 | 0.764378 | 0.386835 |
| ABA 1h | 12.8156 | 4.61616 | 0.564293 | 0.18763 |
| ACC 1h | 27.3096 | 11.1493 | 0.951241 | 0.458854 |
| BABA 1h | 22.1599 | 3.64802 | 0.996566 | 0.16939 |
| Chitin 1h | 13.691 | 4.48051 | 0.594147 | 0.286566 |
| Epi 1h | 14.6002 | 3.94905 | 0.678509 | 0.261346 |
| SA 1h | 94.7185 | 23.7739 | 3.84859 | 0.980422 |
| Me-JA 1h | 239.267 | 14.2058 | 12.953 | 1.05354 |
| Control 6h | 31.9457 | 19.183 | 0.914247 | 0.733676 |
| ABA 6h | 12.6816 | 6.90508 | 0.39888 | 0.215645 |
| ACC 6h | 20.4919 | 12.2451 | 0.558637 | 0.522848 |
| BABA 6h | 49.5144 | 7.7158 | 1.90734 | 0.292651 |
| Chitin 6h | 18.24 | 5.64461 | 0.676821 | 0.288378 |
| Epi 6h | 17.9736 | 9.07873 | 0.555901 | 0.302865 |
| SA 6h | 40.3925 | 9.73388 | 1.67492 | 0.669856 |
| Me-JA 6h | 100.962 | 7.42531 | 4.45565 | 0.51265 |
Source Transcript PGSC0003DMT400008555 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G10600.1 | +3 | 4e-135 | 407 | 220/506 (43%) | cytochrome P450, family 81, subfamily K, polypeptide 2 | chr5:3351227-3352777 FORWARD LENGTH=516 |
