Probe CUST_5090_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5090_PI426222305 | JHI_St_60k_v1 | DMT400009233 | GATTGGATGAAGTTGATGAGGCCACTAAATGAAGAGAAAGATCACTGGGTTCCCGATGAA |
All Microarray Probes Designed to Gene DMG400003585
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5090_PI426222305 | JHI_St_60k_v1 | DMT400009233 | GATTGGATGAAGTTGATGAGGCCACTAAATGAAGAGAAAGATCACTGGGTTCCCGATGAA |
| CUST_5277_PI426222305 | JHI_St_60k_v1 | DMT400009232 | AGTTTGGCAACTCATCTATTTTCTACTATCCTCGTTTTTCTCATACAGGTTCCCGATGAA |
Microarray Signals from CUST_5090_PI426222305
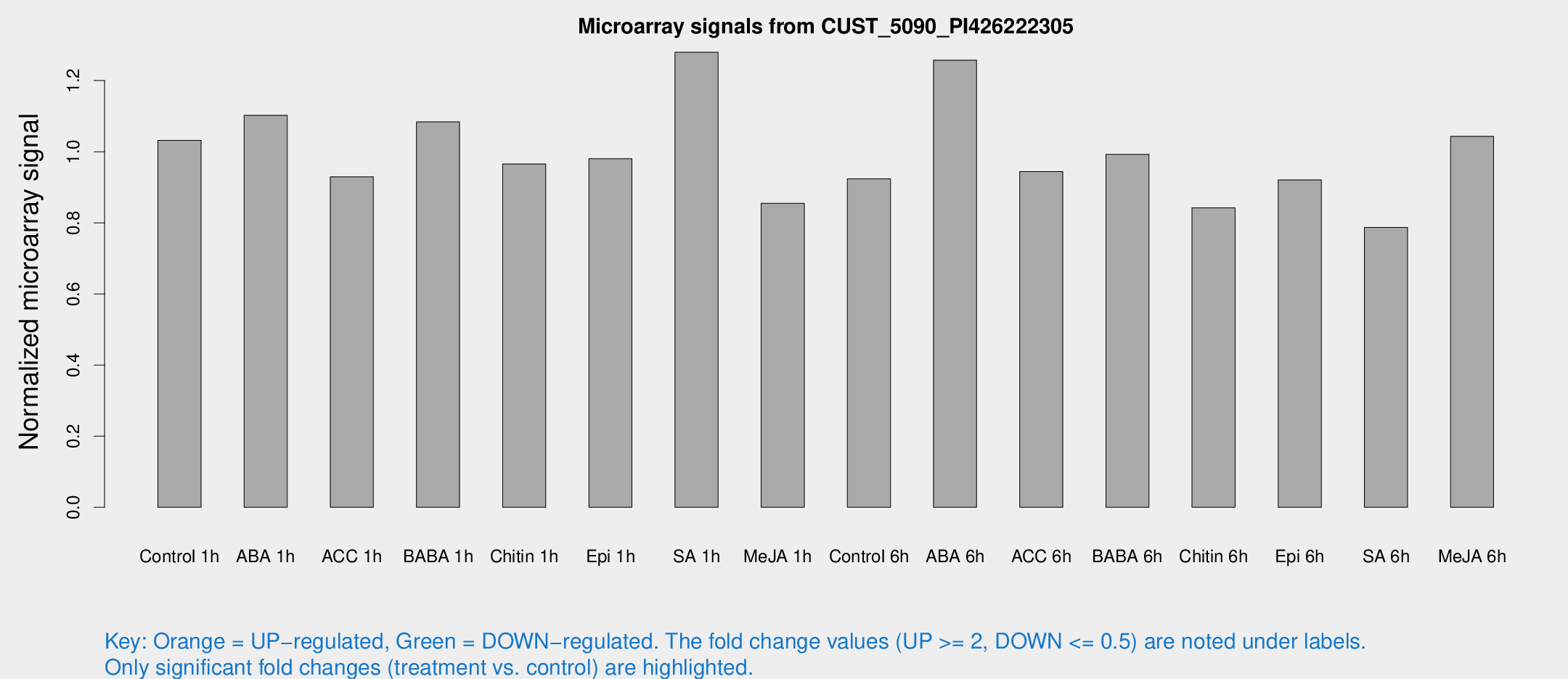
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 754.449 | 119.819 | 1.03169 | 0.0961436 |
| ABA 1h | 703.801 | 68.991 | 1.10234 | 0.0637956 |
| ACC 1h | 726.715 | 163.529 | 0.929499 | 0.179183 |
| BABA 1h | 774.381 | 142.194 | 1.08391 | 0.110178 |
| Chitin 1h | 630.901 | 85.742 | 0.965379 | 0.066528 |
| Epi 1h | 608.559 | 38.4369 | 0.980172 | 0.0567779 |
| SA 1h | 955.745 | 122.099 | 1.27977 | 0.0959693 |
| Me-JA 1h | 501.504 | 37.5033 | 0.855053 | 0.0496136 |
| Control 6h | 712.56 | 174.911 | 0.923602 | 0.189249 |
| ABA 6h | 964.9 | 97.6366 | 1.25738 | 0.0727067 |
| ACC 6h | 787.713 | 104.105 | 0.944445 | 0.0546875 |
| BABA 6h | 801.533 | 77.4424 | 0.992646 | 0.0587263 |
| Chitin 6h | 640.716 | 37.1455 | 0.842291 | 0.0488161 |
| Epi 6h | 746.469 | 60.841 | 0.920478 | 0.0587557 |
| SA 6h | 607.868 | 156.728 | 0.787286 | 0.160053 |
| Me-JA 6h | 751.007 | 80.1466 | 1.04315 | 0.0603694 |
Source Transcript PGSC0003DMT400009233 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G20110.1 | +3 | 3e-169 | 504 | 300/604 (50%) | RING/FYVE/PHD zinc finger superfamily protein | chr1:6971554-6974578 FORWARD LENGTH=601 |
