Probe CUST_50839_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50839_PI426222305 | JHI_St_60k_v1 | DMT400080660 | CCTATTTGGCTTCCCTCTATTTCGTCAAGTATACTAAGCTAGTTTTGATCATAGATTTTC |
All Microarray Probes Designed to Gene DMG400031411
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50839_PI426222305 | JHI_St_60k_v1 | DMT400080660 | CCTATTTGGCTTCCCTCTATTTCGTCAAGTATACTAAGCTAGTTTTGATCATAGATTTTC |
| CUST_50845_PI426222305 | JHI_St_60k_v1 | DMT400080659 | CGTATTCATCTTTTCTTAATGTGCTCACACTTATGGCTCTTACACACCATCTTGTTCACC |
Microarray Signals from CUST_50839_PI426222305
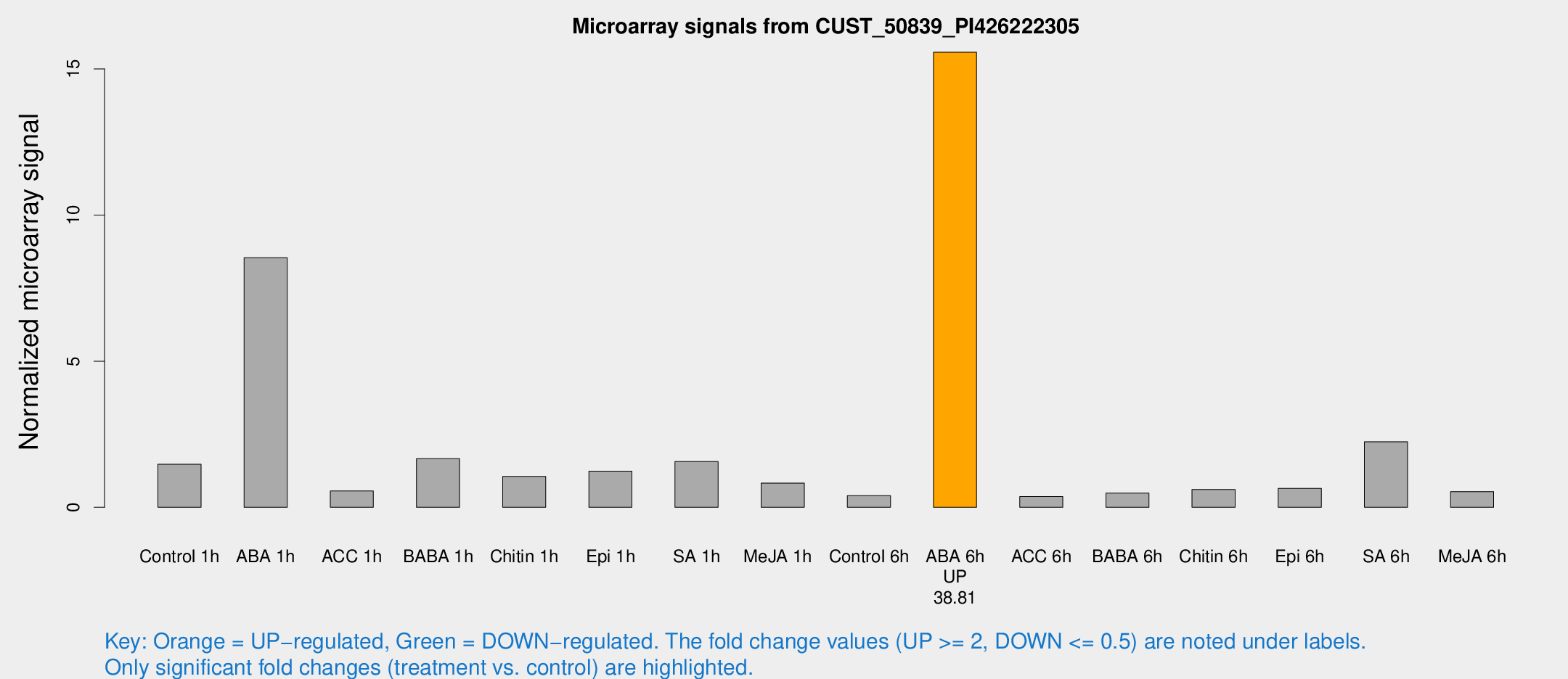
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 59.8377 | 5.27607 | 1.47364 | 0.23292 |
| ABA 1h | 332.106 | 85.3204 | 8.54133 | 2.35941 |
| ACC 1h | 27.2728 | 8.90952 | 0.559475 | 0.228129 |
| BABA 1h | 65.7746 | 9.2736 | 1.66337 | 0.184932 |
| Chitin 1h | 41.1691 | 11.3036 | 1.05391 | 0.264843 |
| Epi 1h | 50.9418 | 18.1309 | 1.2388 | 0.60657 |
| SA 1h | 66.4569 | 10.8381 | 1.56749 | 0.165428 |
| Me-JA 1h | 29.5228 | 8.74989 | 0.829644 | 0.171314 |
| Control 6h | 19.1438 | 6.29545 | 0.401224 | 0.258351 |
| ABA 6h | 669.048 | 40.3366 | 15.5706 | 0.903734 |
| ACC 6h | 17.4255 | 4.66016 | 0.366433 | 0.12362 |
| BABA 6h | 26.2546 | 10.6652 | 0.48418 | 0.221215 |
| Chitin 6h | 29.771 | 9.03278 | 0.610464 | 0.292726 |
| Epi 6h | 51.4722 | 36.6887 | 0.649217 | 0.62951 |
| SA 6h | 150.284 | 101.86 | 2.24382 | 2.03555 |
| Me-JA 6h | 22.5797 | 4.6015 | 0.536053 | 0.108447 |
Source Transcript PGSC0003DMT400080660 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G22600.1 | +3 | 2e-45 | 166 | 118/354 (33%) | Late embryogenesis abundant protein (LEA) family protein | chr1:7987308-7989187 REVERSE LENGTH=385 |
