Probe CUST_5036_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5036_PI426222305 | JHI_St_60k_v1 | DMT400009066 | AGTGTTATTGGCTTCATTGATCCTTTTCTTAGTGCTATTCAATGTTATATGCAGCTAGTG |
All Microarray Probes Designed to Gene DMG400003528
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5036_PI426222305 | JHI_St_60k_v1 | DMT400009066 | AGTGTTATTGGCTTCATTGATCCTTTTCTTAGTGCTATTCAATGTTATATGCAGCTAGTG |
| CUST_5363_PI426222305 | JHI_St_60k_v1 | DMT400009067 | GCTCGAGTGTTATTGGCTTCATTGATCCTTTTCTTAGTGCTATTCAATGTTATATGCAGC |
Microarray Signals from CUST_5036_PI426222305
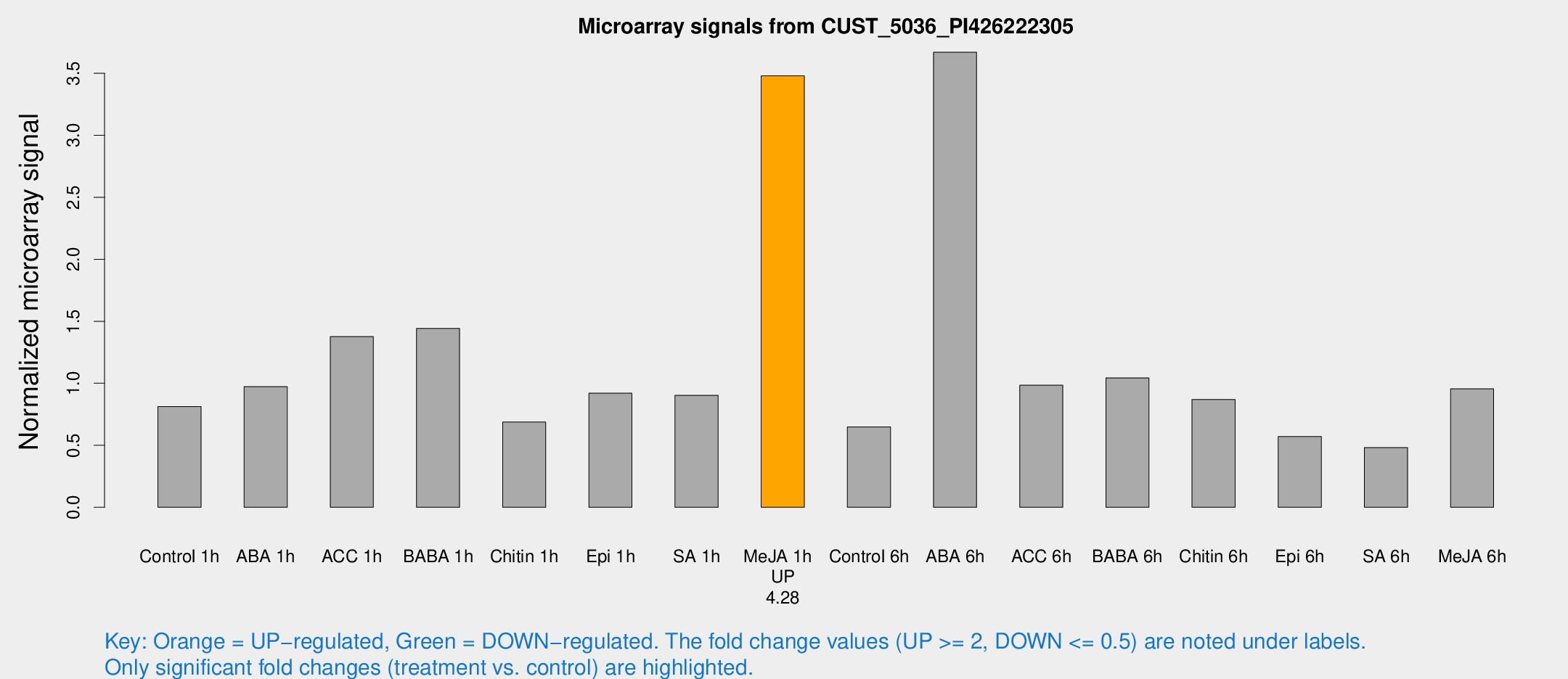
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.0346 | 7.42184 | 0.812427 | 0.180945 |
| ABA 1h | 27.6381 | 8.31436 | 0.972902 | 0.343083 |
| ACC 1h | 51.5829 | 20.9641 | 1.3764 | 0.785039 |
| BABA 1h | 40.0566 | 4.38717 | 1.44304 | 0.194468 |
| Chitin 1h | 18.5777 | 4.27269 | 0.687777 | 0.147174 |
| Epi 1h | 23.3079 | 4.0573 | 0.919864 | 0.161955 |
| SA 1h | 28.9231 | 8.92867 | 0.902839 | 0.270155 |
| Me-JA 1h | 81.3295 | 6.11109 | 3.47899 | 0.243548 |
| Control 6h | 19.5552 | 4.32049 | 0.648069 | 0.135961 |
| ABA 6h | 116.241 | 22.6395 | 3.66954 | 0.683889 |
| ACC 6h | 33.1549 | 5.94989 | 0.984037 | 0.130173 |
| BABA 6h | 33.5701 | 4.11386 | 1.04349 | 0.129032 |
| Chitin 6h | 26.5061 | 3.9369 | 0.869275 | 0.131689 |
| Epi 6h | 20.2528 | 6.71789 | 0.570354 | 0.180074 |
| SA 6h | 15.6088 | 4.85239 | 0.481493 | 0.162165 |
| Me-JA 6h | 27.9535 | 5.19094 | 0.955221 | 0.131321 |
Source Transcript PGSC0003DMT400009066 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G18170.1 | +1 | 0.0 | 629 | 294/367 (80%) | MAP kinase 7 | chr2:7908178-7909374 REVERSE LENGTH=368 |
