Probe CUST_50341_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50341_PI426222305 | JHI_St_60k_v1 | DMT400045838 | CGGACTCTGCAAAAATGTTGTCGTAGTATATGTAGGTGACATTTTTAAAGAGTTTGAAAG |
All Microarray Probes Designed to Gene DMG400017779
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_50341_PI426222305 | JHI_St_60k_v1 | DMT400045838 | CGGACTCTGCAAAAATGTTGTCGTAGTATATGTAGGTGACATTTTTAAAGAGTTTGAAAG |
| CUST_50344_PI426222305 | JHI_St_60k_v1 | DMT400045837 | AACTCATGGATGACAGTCTAGTAGATTGGATTTTACGTGCCTTGGATGGCGATTCATCTT |
Microarray Signals from CUST_50341_PI426222305
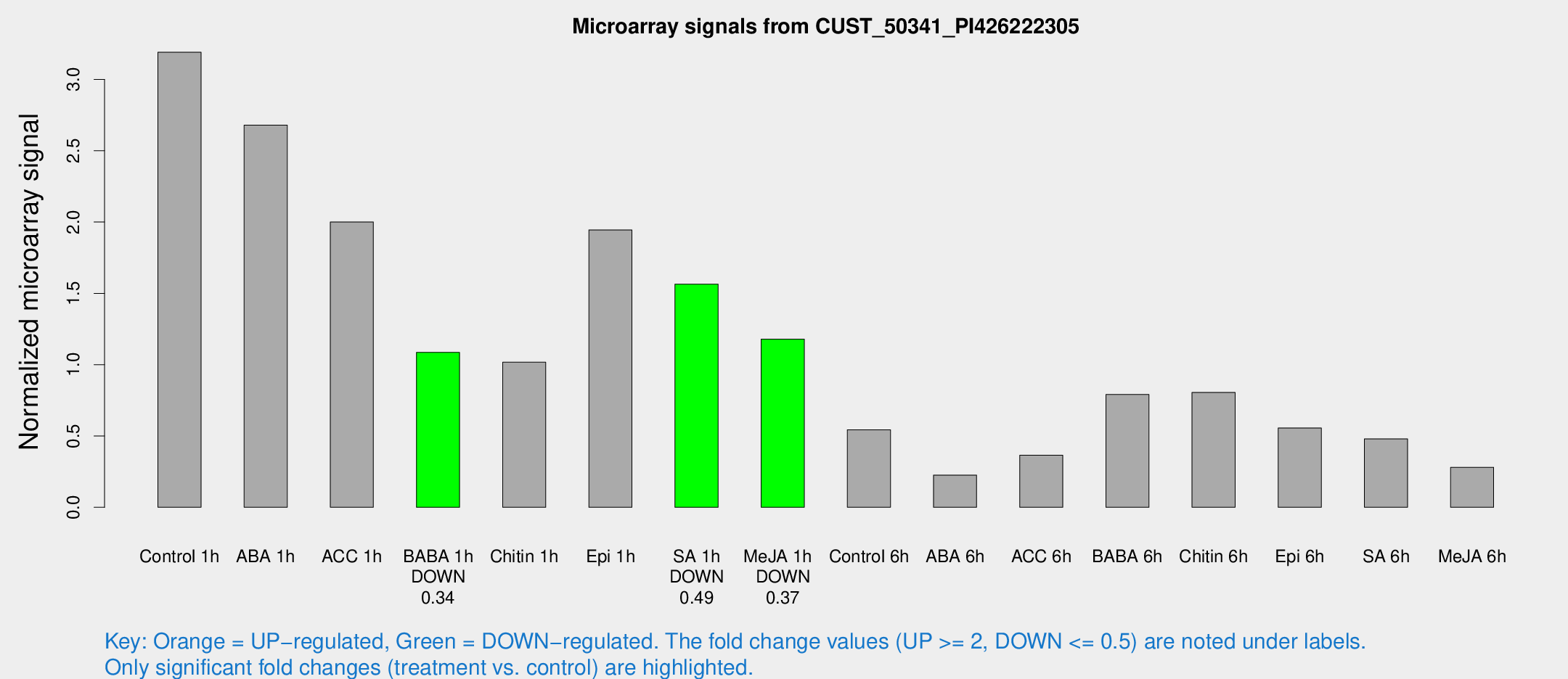
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 99.9032 | 19.1059 | 3.19084 | 0.383799 |
| ABA 1h | 72.1922 | 6.05202 | 2.67943 | 0.193056 |
| ACC 1h | 62.3705 | 5.00645 | 2.00061 | 0.202844 |
| BABA 1h | 32.9872 | 6.29327 | 1.08615 | 0.137012 |
| Chitin 1h | 31.0805 | 10.3971 | 1.01771 | 0.443154 |
| Epi 1h | 53.7935 | 13.1919 | 1.94459 | 0.411227 |
| SA 1h | 48.7514 | 4.20873 | 1.56427 | 0.146361 |
| Me-JA 1h | 30.2062 | 6.04991 | 1.17928 | 0.150294 |
| Control 6h | 18.0264 | 4.6883 | 0.54398 | 0.141451 |
| ABA 6h | 7.55883 | 3.50667 | 0.225956 | 0.113103 |
| ACC 6h | 13.6937 | 4.12814 | 0.365347 | 0.116145 |
| BABA 6h | 38.23 | 16.4857 | 0.790909 | 0.745432 |
| Chitin 6h | 26.8473 | 4.61373 | 0.805519 | 0.161162 |
| Epi 6h | 19.2531 | 4.02769 | 0.556181 | 0.126186 |
| SA 6h | 14.4141 | 3.62514 | 0.479703 | 0.122186 |
| Me-JA 6h | 10.0252 | 4.35473 | 0.280392 | 0.129706 |
Source Transcript PGSC0003DMT400045838 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G34440.1 | +1 | 1e-166 | 503 | 281/467 (60%) | Protein kinase superfamily protein | chr4:16466008-16468748 FORWARD LENGTH=670 |
