Probe CUST_49441_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49441_PI426222305 | JHI_St_60k_v1 | DMT400051191 | TTAAATAAATCTTTCTTTAACCGCAGGATGCAGACCTATACACGGAAAGGATCTTCCAGA |
All Microarray Probes Designed to Gene DMG400019882
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49430_PI426222305 | JHI_St_60k_v1 | DMT400051190 | AAAGACTTGCAAGAGCCTGTTCGTATTCCAGGATGCAGACCTATACACGGAAAGGATCTT |
| CUST_49441_PI426222305 | JHI_St_60k_v1 | DMT400051191 | TTAAATAAATCTTTCTTTAACCGCAGGATGCAGACCTATACACGGAAAGGATCTTCCAGA |
Microarray Signals from CUST_49441_PI426222305
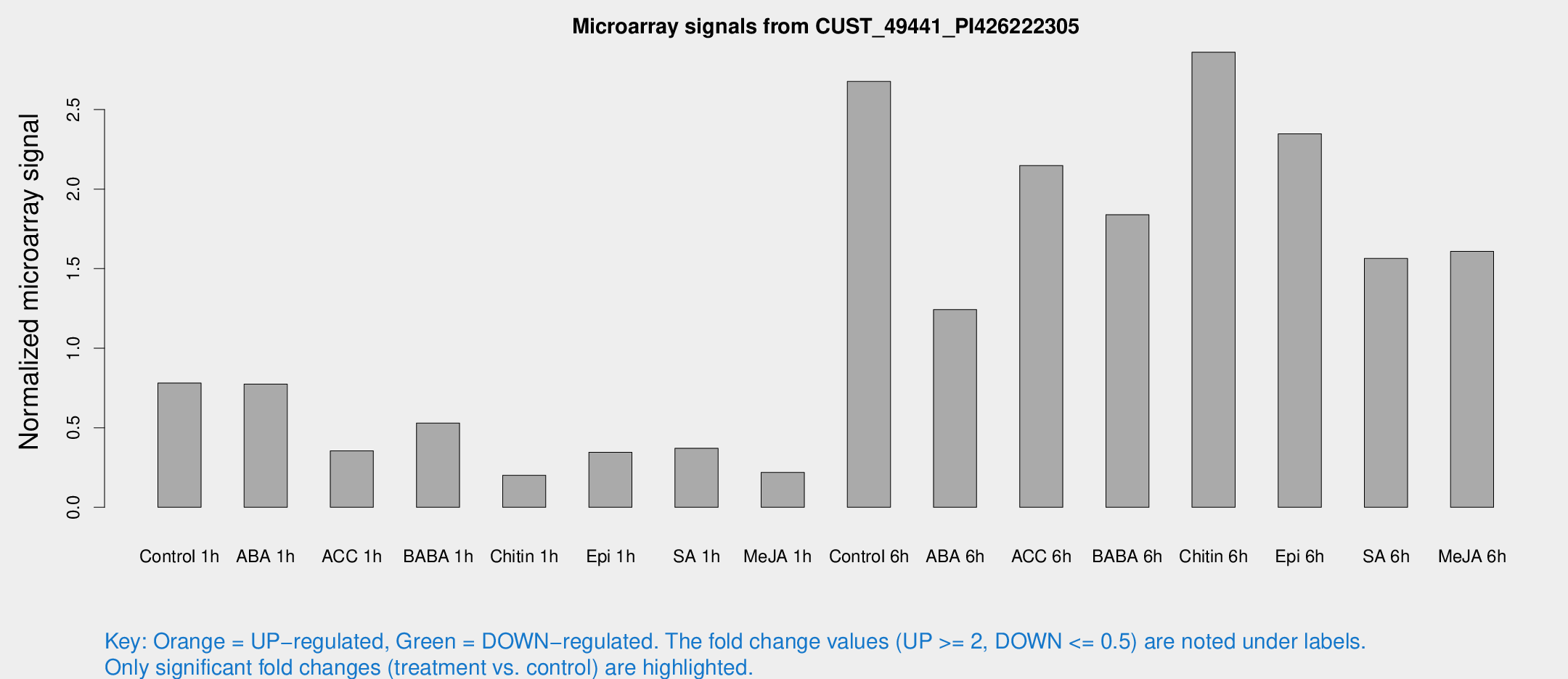
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 40.6655 | 4.77853 | 0.781108 | 0.115093 |
| ABA 1h | 35.9896 | 5.31671 | 0.774081 | 0.180035 |
| ACC 1h | 20.782 | 6.07267 | 0.355234 | 0.116778 |
| BABA 1h | 27.3841 | 5.30217 | 0.529672 | 0.0894087 |
| Chitin 1h | 10.7448 | 4.28785 | 0.201535 | 0.0878733 |
| Epi 1h | 17.7624 | 5.60419 | 0.345798 | 0.156357 |
| SA 1h | 23.6437 | 8.2938 | 0.371154 | 0.223643 |
| Me-JA 1h | 9.27925 | 3.87342 | 0.219437 | 0.0950244 |
| Control 6h | 145.813 | 32.469 | 2.67669 | 0.45338 |
| ABA 6h | 69.8207 | 12.212 | 1.24257 | 0.147857 |
| ACC 6h | 130.679 | 24.7688 | 2.14844 | 0.144122 |
| BABA 6h | 110.865 | 24.4922 | 1.839 | 0.376257 |
| Chitin 6h | 158.626 | 19.0192 | 2.86032 | 0.386189 |
| Epi 6h | 143.376 | 31.68 | 2.34791 | 0.727206 |
| SA 6h | 81.6004 | 13.7223 | 1.56416 | 0.467586 |
| Me-JA 6h | 93.618 | 31.6701 | 1.60818 | 0.49578 |
Source Transcript PGSC0003DMT400051191 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G01070.1 | +3 | 2e-119 | 357 | 177/297 (60%) | UDP-Glycosyltransferase superfamily protein | chr4:461858-463300 REVERSE LENGTH=480 |
