Probe CUST_48981_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48981_PI426222305 | JHI_St_60k_v1 | DMT400056252 | GCTCATATTAGTGGTGAATTGTGTCCTATTTTGAAAAATGCAGGGGCCTTGTGGCTATGA |
All Microarray Probes Designed to Gene DMG400021856
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48981_PI426222305 | JHI_St_60k_v1 | DMT400056252 | GCTCATATTAGTGGTGAATTGTGTCCTATTTTGAAAAATGCAGGGGCCTTGTGGCTATGA |
| CUST_48986_PI426222305 | JHI_St_60k_v1 | DMT400056253 | ACGTACCTCTAGGTGAACTCATGGCTTGTCTTGAAATACAATATTATTACTTTCTCTGAT |
Microarray Signals from CUST_48981_PI426222305
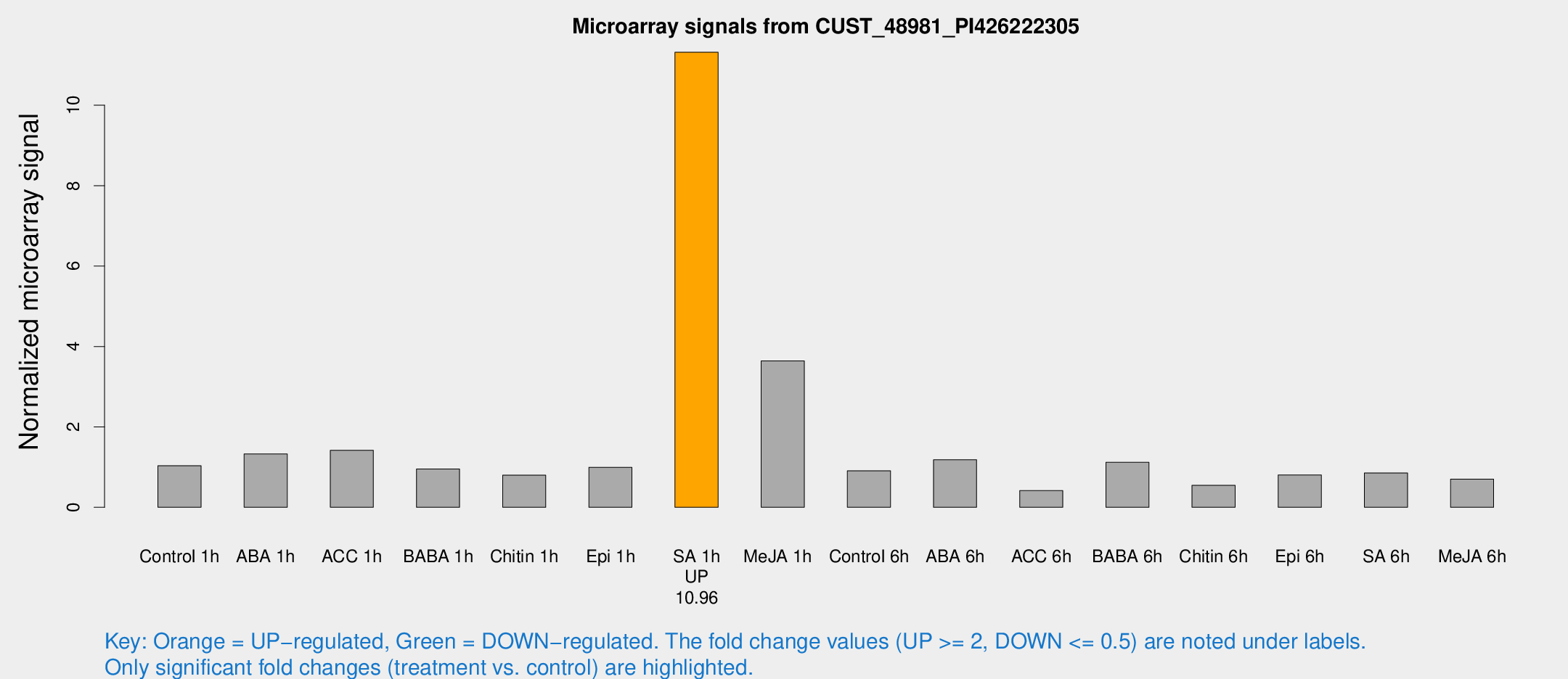
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 51.3873 | 20.3217 | 1.03269 | 0.422456 |
| ABA 1h | 48.9975 | 4.40105 | 1.3272 | 0.119128 |
| ACC 1h | 61.5981 | 7.32043 | 1.41922 | 0.205518 |
| BABA 1h | 44.3367 | 15.2437 | 0.952332 | 0.313018 |
| Chitin 1h | 30.5915 | 4.10887 | 0.799863 | 0.107534 |
| Epi 1h | 37.8085 | 8.62702 | 0.995103 | 0.207748 |
| SA 1h | 502.622 | 92.4619 | 11.3191 | 1.863 |
| Me-JA 1h | 129.718 | 29.1664 | 3.64066 | 0.516691 |
| Control 6h | 42.221 | 12.2987 | 0.906529 | 0.241346 |
| ABA 6h | 55.4534 | 11.9495 | 1.18117 | 0.314938 |
| ACC 6h | 20.4645 | 4.73211 | 0.416319 | 0.0949842 |
| BABA 6h | 60.0363 | 18.7602 | 1.11878 | 0.525704 |
| Chitin 6h | 24.2936 | 4.26329 | 0.545363 | 0.0963499 |
| Epi 6h | 38.4943 | 5.43093 | 0.802296 | 0.160853 |
| SA 6h | 35.2625 | 4.37193 | 0.852344 | 0.106553 |
| Me-JA 6h | 30.2303 | 6.05259 | 0.700208 | 0.101643 |
Source Transcript PGSC0003DMT400056252 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G28480.1 | +1 | 7e-38 | 127 | 63/104 (61%) | Thioredoxin superfamily protein | chr1:10013634-10014047 REVERSE LENGTH=137 |
