Probe CUST_48526_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48526_PI426222305 | JHI_St_60k_v1 | DMT400043382 | GGAAGCATCCTAAGTCATGTAAATGTTTGTCTTTTGATTGTCACAATCTGAACAAACATC |
All Microarray Probes Designed to Gene DMG400016842
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_48526_PI426222305 | JHI_St_60k_v1 | DMT400043382 | GGAAGCATCCTAAGTCATGTAAATGTTTGTCTTTTGATTGTCACAATCTGAACAAACATC |
| CUST_48523_PI426222305 | JHI_St_60k_v1 | DMT400043381 | GGAAGCATCCTAAGTCATGTAAATGTTTGTCTTTTGATTGTCACAATCTGAACAAACATC |
Microarray Signals from CUST_48526_PI426222305
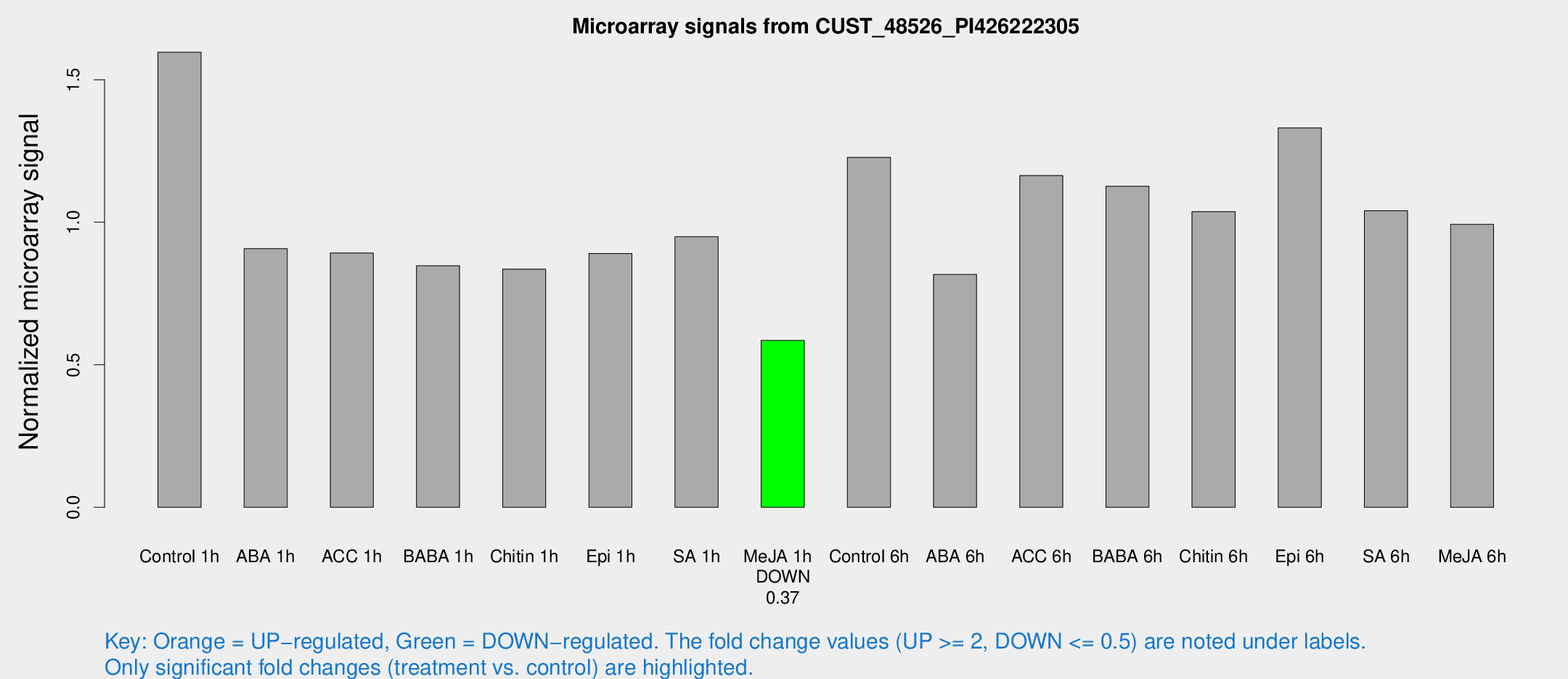
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 281.149 | 63.3206 | 1.59639 | 0.242919 |
| ABA 1h | 135.607 | 10.0303 | 0.907241 | 0.134022 |
| ACC 1h | 161.456 | 32.0867 | 0.892301 | 0.136476 |
| BABA 1h | 144.202 | 29.0116 | 0.847708 | 0.107875 |
| Chitin 1h | 126.514 | 7.96984 | 0.835581 | 0.0525209 |
| Epi 1h | 131.343 | 15.71 | 0.890206 | 0.0867548 |
| SA 1h | 168.726 | 29.6492 | 0.949618 | 0.125723 |
| Me-JA 1h | 80.632 | 5.67207 | 0.585203 | 0.0410238 |
| Control 6h | 216.264 | 42.2451 | 1.22713 | 0.163619 |
| ABA 6h | 148.095 | 17.2775 | 0.816758 | 0.0724769 |
| ACC 6h | 228.837 | 33.3017 | 1.16369 | 0.0699889 |
| BABA 6h | 212.621 | 12.866 | 1.1265 | 0.0843898 |
| Chitin 6h | 185.709 | 11.3211 | 1.03723 | 0.063182 |
| Epi 6h | 257.783 | 39.5123 | 1.33119 | 0.198032 |
| SA 6h | 183.513 | 41.0361 | 1.04059 | 0.145415 |
| Me-JA 6h | 166.916 | 11.5598 | 0.992621 | 0.0606635 |
Source Transcript PGSC0003DMT400043382 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G32910.1 | +3 | 1e-46 | 179 | 105/236 (44%) | DCD (Development and Cell Death) domain protein | chr2:13959685-13961989 FORWARD LENGTH=691 |
