Probe CUST_47445_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47445_PI426222305 | JHI_St_60k_v1 | DMT400064776 | GTTTCCCTTCATTACTCTTTGGTTTGTTCAGGGGAGATTTTGGAAAATGGCTATCTTTGA |
All Microarray Probes Designed to Gene DMG400025160
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_47466_PI426222305 | JHI_St_60k_v1 | DMT400064777 | GGCACTAGTTCATCGTTAATTTTGACAATTGTGAGTGCAGATTGGCTTGAAGCACCAGCA |
| CUST_47445_PI426222305 | JHI_St_60k_v1 | DMT400064776 | GTTTCCCTTCATTACTCTTTGGTTTGTTCAGGGGAGATTTTGGAAAATGGCTATCTTTGA |
Microarray Signals from CUST_47445_PI426222305
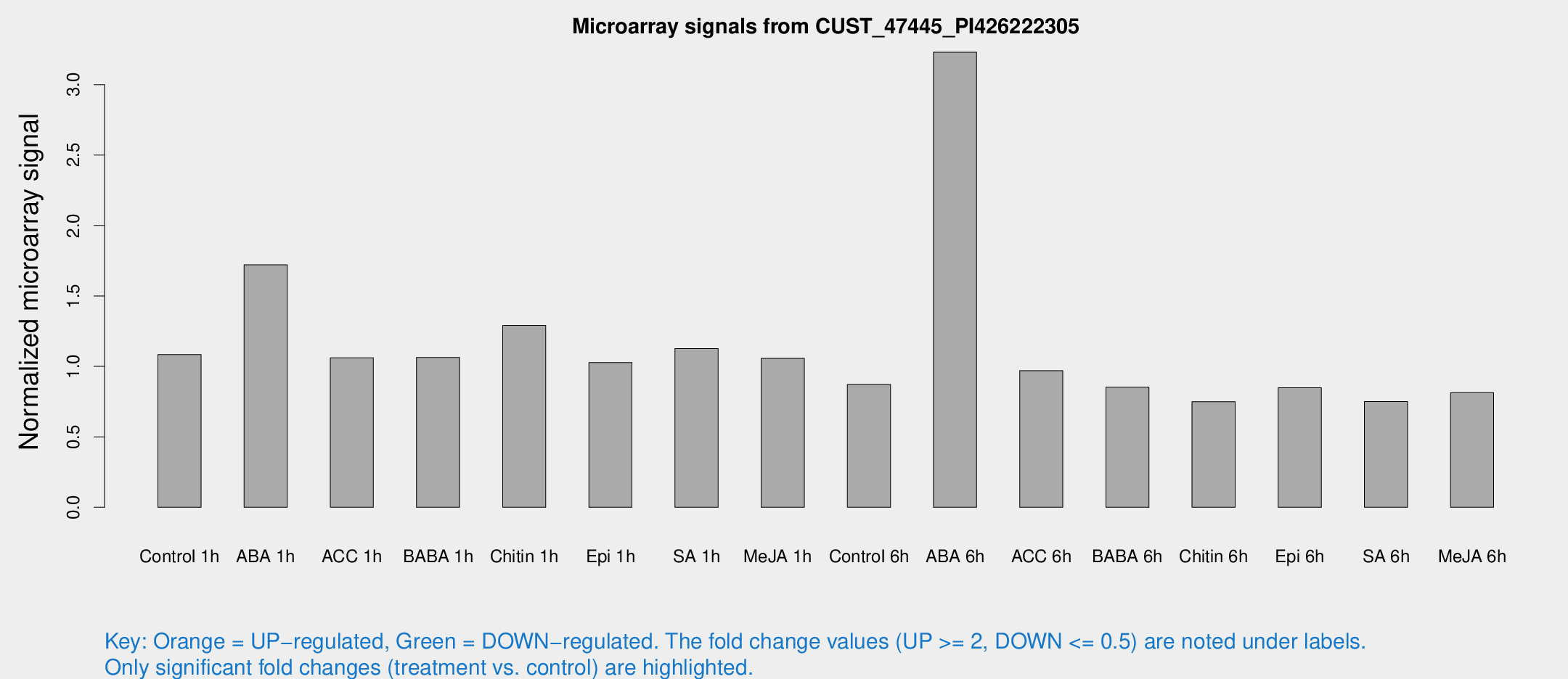
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6418.55 | 418.069 | 1.08354 | 0.0625601 |
| ABA 1h | 9079.65 | 927.512 | 1.72111 | 0.13537 |
| ACC 1h | 6469.7 | 508.742 | 1.06122 | 0.0612726 |
| BABA 1h | 6401.04 | 1412.79 | 1.06349 | 0.158136 |
| Chitin 1h | 6913.96 | 664.018 | 1.29077 | 0.0745245 |
| Epi 1h | 5274.15 | 406.347 | 1.02697 | 0.0784322 |
| SA 1h | 7007.88 | 1078.78 | 1.12624 | 0.125093 |
| Me-JA 1h | 5183.03 | 697.043 | 1.05659 | 0.122848 |
| Control 6h | 5549.38 | 1423.67 | 0.871303 | 0.182526 |
| ABA 6h | 21371.5 | 4631.89 | 3.22977 | 0.619857 |
| ACC 6h | 6628.3 | 676.81 | 0.970026 | 0.056007 |
| BABA 6h | 5635.08 | 325.834 | 0.85229 | 0.0492097 |
| Chitin 6h | 4715.26 | 272.752 | 0.7497 | 0.0533548 |
| Epi 6h | 5747.72 | 815.379 | 0.848086 | 0.115938 |
| SA 6h | 4648.34 | 1007.43 | 0.751433 | 0.102419 |
| Me-JA 6h | 4825.74 | 473.169 | 0.814132 | 0.0470069 |
Source Transcript PGSC0003DMT400064776 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50830.1 | +3 | 2e-86 | 263 | 134/201 (67%) | cold-regulated 413-plasma membrane 2 | chr3:18894109-18895355 REVERSE LENGTH=203 |
