Probe CUST_46296_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46296_PI426222305 | JHI_St_60k_v1 | DMT400034138 | CCTTTGGACAGGTAAACAGACAGTTCAATTTAGTCACAATTTATACATACTCTGGAGTTA |
All Microarray Probes Designed to Gene DMG401013112
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46296_PI426222305 | JHI_St_60k_v1 | DMT400034138 | CCTTTGGACAGGTAAACAGACAGTTCAATTTAGTCACAATTTATACATACTCTGGAGTTA |
| CUST_46305_PI426222305 | JHI_St_60k_v1 | DMT400034124 | CCAGAGAAGATAAAAGACACAACAATTGTGTATATGTCACAACTCACAACTTGAAGACTT |
Microarray Signals from CUST_46296_PI426222305
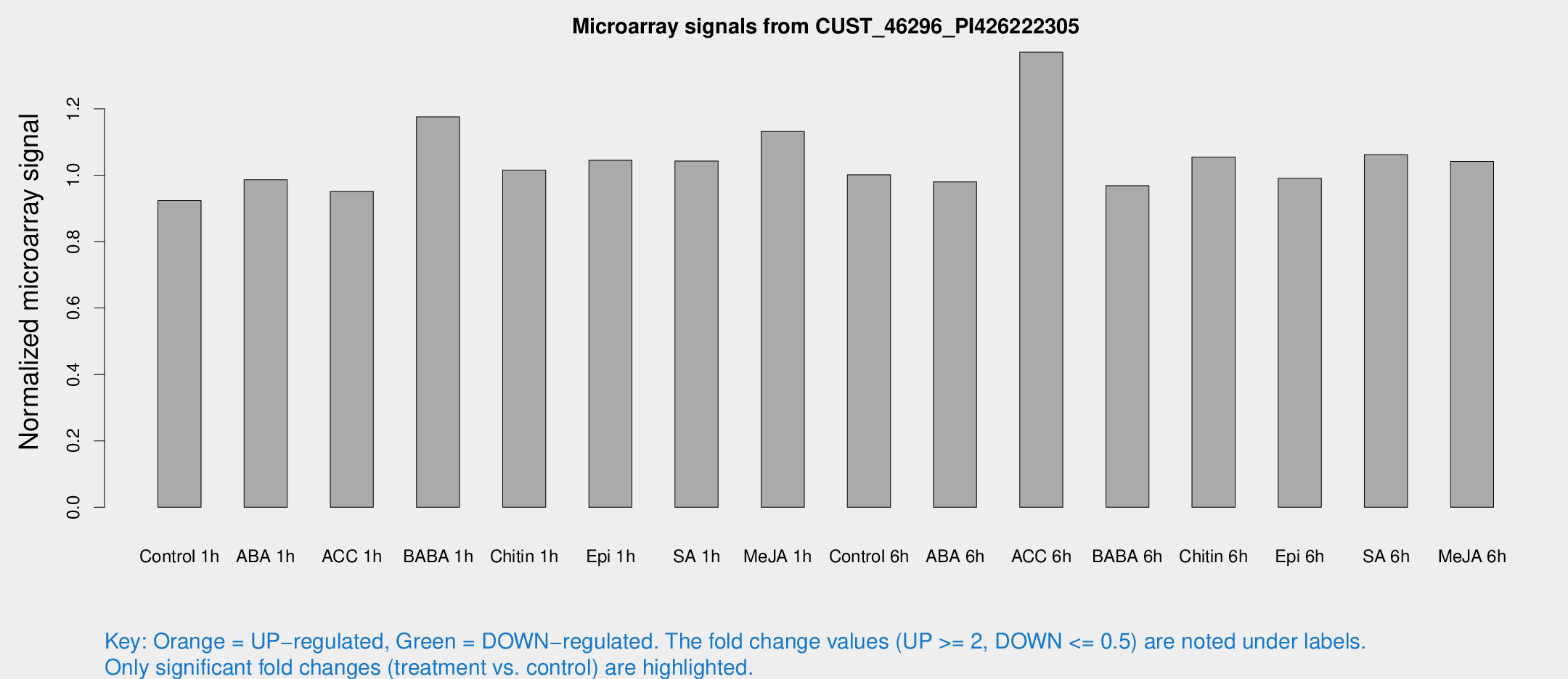
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.56828 | 3.22683 | 0.923369 | 0.534679 |
| ABA 1h | 5.25819 | 3.04535 | 0.986332 | 0.571176 |
| ACC 1h | 5.89063 | 3.41267 | 0.95147 | 0.550972 |
| BABA 1h | 7.06676 | 3.41201 | 1.17557 | 0.600794 |
| Chitin 1h | 5.50909 | 3.21027 | 1.01535 | 0.585312 |
| Epi 1h | 5.45808 | 3.16926 | 1.045 | 0.605952 |
| SA 1h | 6.69556 | 3.11353 | 1.04245 | 0.521285 |
| Me-JA 1h | 5.59061 | 3.25216 | 1.13138 | 0.655108 |
| Control 6h | 6.05457 | 3.51058 | 1.00123 | 0.57981 |
| ABA 6h | 6.28081 | 3.63887 | 0.979874 | 0.567406 |
| ACC 6h | 10.3587 | 4.1385 | 1.37019 | 0.645427 |
| BABA 6h | 6.56027 | 3.81634 | 0.96842 | 0.560893 |
| Chitin 6h | 6.76829 | 3.92312 | 1.05443 | 0.610749 |
| Epi 6h | 6.81204 | 3.99833 | 0.990557 | 0.574282 |
| SA 6h | 6.33008 | 3.66973 | 1.06172 | 0.61534 |
| Me-JA 6h | 6.26458 | 3.45885 | 1.04137 | 0.575715 |
Source Transcript PGSC0003DMT400034138 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G15520.1 | +2 | 1e-118 | 191 | 87/117 (74%) | pleiotropic drug resistance 12 | chr1:5331993-5338175 REVERSE LENGTH=1423 |
