Probe CUST_45938_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45938_PI426222305 | JHI_St_60k_v1 | DMT400048002 | TTTTGCTTTCAGAGGCCACAATCTCATACTTGAGATTCAGGCGACAATGCCTTCGAGTGA |
All Microarray Probes Designed to Gene DMG400018651
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45938_PI426222305 | JHI_St_60k_v1 | DMT400048002 | TTTTGCTTTCAGAGGCCACAATCTCATACTTGAGATTCAGGCGACAATGCCTTCGAGTGA |
| CUST_45890_PI426222305 | JHI_St_60k_v1 | DMT400048001 | GACACTGGTGTTAAAATTAGCTTCTTCAATCCTCAGTAACTAGTATGAACATGCGAAATT |
Microarray Signals from CUST_45938_PI426222305
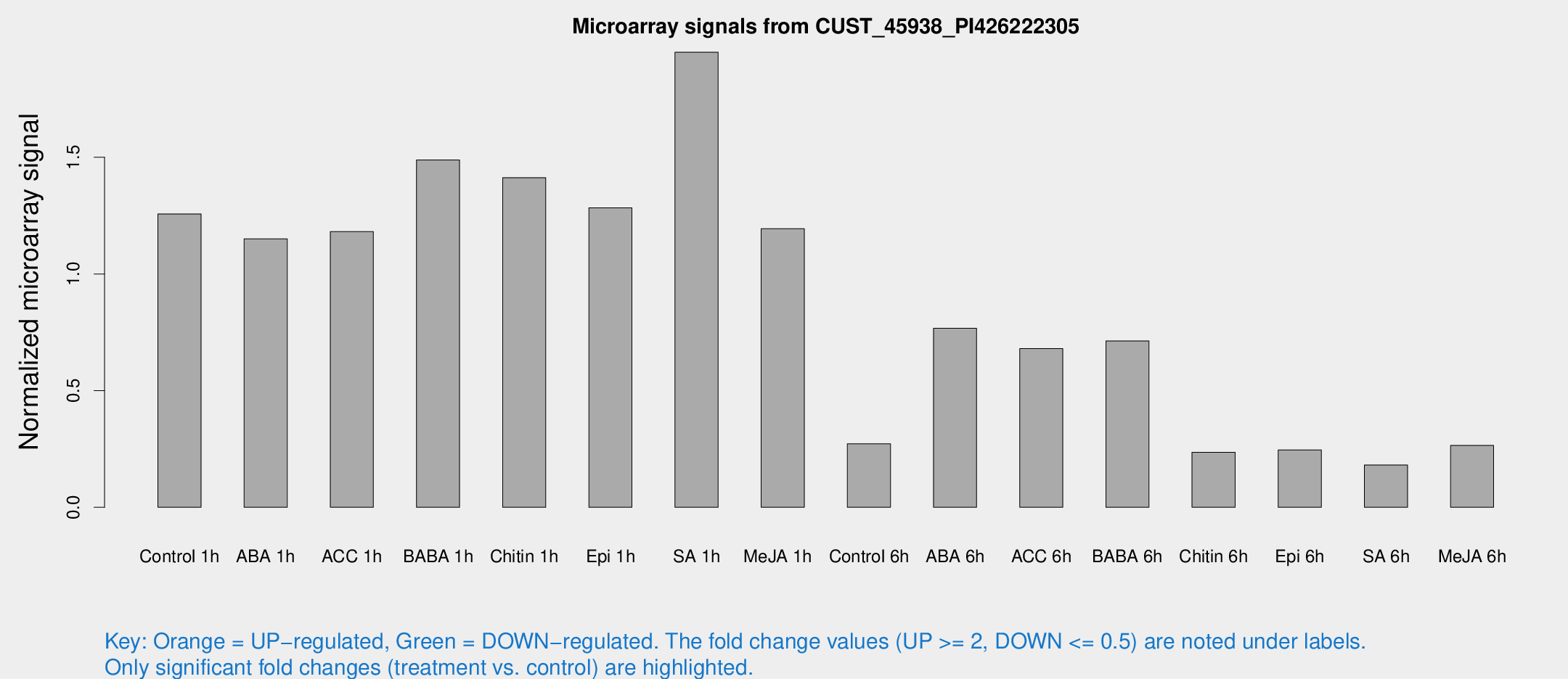
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2732.46 | 350.433 | 1.25737 | 0.0937073 |
| ABA 1h | 2244.57 | 402.629 | 1.15078 | 0.127471 |
| ACC 1h | 3123.02 | 1077.45 | 1.18152 | 0.533638 |
| BABA 1h | 3422.97 | 1017.04 | 1.48856 | 0.39042 |
| Chitin 1h | 2755.95 | 360.643 | 1.41278 | 0.0991484 |
| Epi 1h | 2373.24 | 137.179 | 1.28331 | 0.0741181 |
| SA 1h | 4526.58 | 1014.45 | 1.95026 | 0.367659 |
| Me-JA 1h | 2142.77 | 365.409 | 1.19432 | 0.0996245 |
| Control 6h | 662.6 | 204.049 | 0.272186 | 0.0862413 |
| ABA 6h | 2033.21 | 654.357 | 0.767012 | 0.330168 |
| ACC 6h | 1697.46 | 238.083 | 0.680401 | 0.0393252 |
| BABA 6h | 1886.81 | 631.128 | 0.712716 | 0.209383 |
| Chitin 6h | 552.243 | 90.5555 | 0.235577 | 0.0435688 |
| Epi 6h | 638.675 | 161.194 | 0.246069 | 0.0594931 |
| SA 6h | 448.301 | 157.187 | 0.181263 | 0.060268 |
| Me-JA 6h | 630.793 | 185.171 | 0.265067 | 0.0770118 |
Source Transcript PGSC0003DMT400048002 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G47670.1 | +3 | 0.0 | 573 | 281/543 (52%) | Transmembrane amino acid transporter family protein | chr1:17536834-17539486 REVERSE LENGTH=519 |
