Probe CUST_45329_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45329_PI426222305 | JHI_St_60k_v1 | DMT400001688 | TTTCCAGTGTGAGAGCATAGATGCAGTGGAGAAGAAGCTGACAGAAATGGGGATAAAATA |
All Microarray Probes Designed to Gene DMG400000631
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45329_PI426222305 | JHI_St_60k_v1 | DMT400001688 | TTTCCAGTGTGAGAGCATAGATGCAGTGGAGAAGAAGCTGACAGAAATGGGGATAAAATA |
| CUST_45322_PI426222305 | JHI_St_60k_v1 | DMT400001687 | GTTAGAGTAGAATGGAATCGAATACATCTTGTAGAATTGAACACTCTGTAAAGTTTCACC |
Microarray Signals from CUST_45329_PI426222305
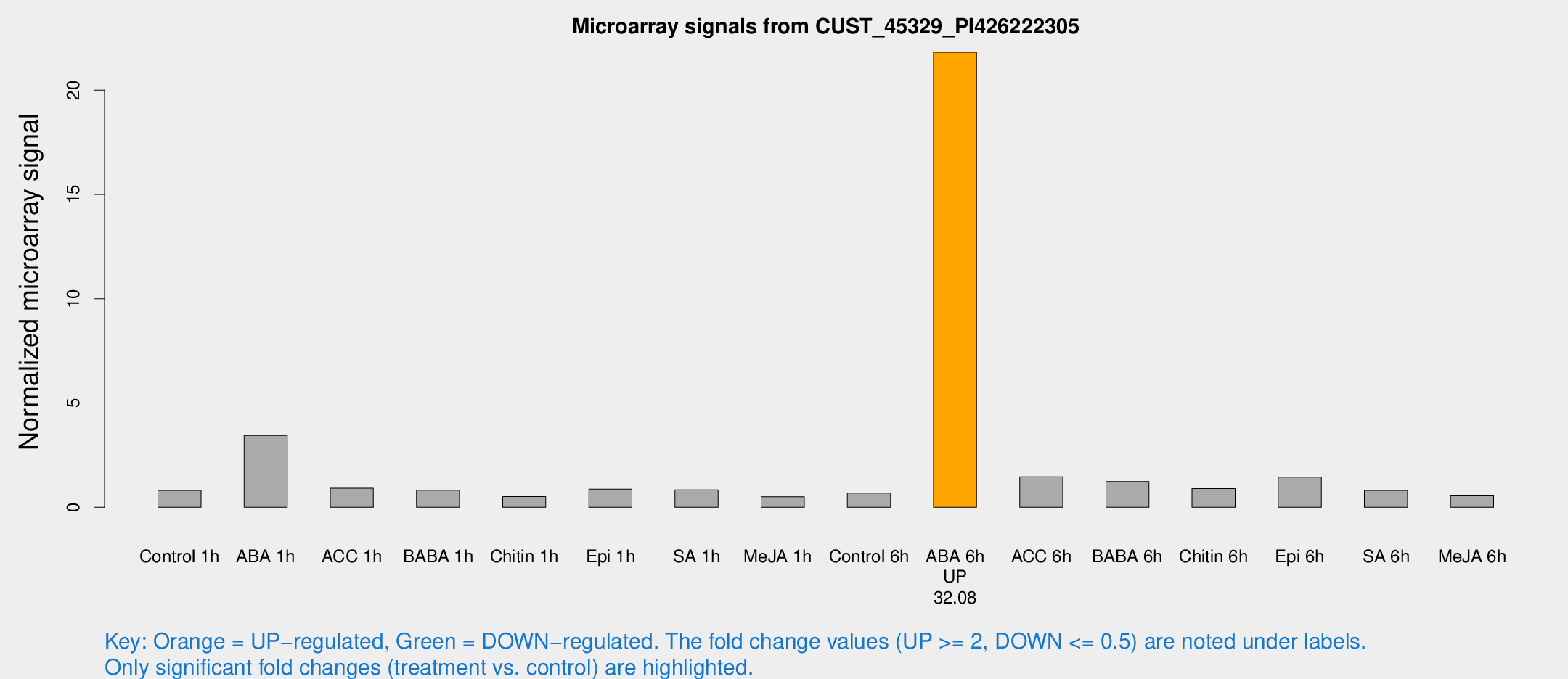
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 39.5283 | 5.47716 | 0.815137 | 0.0807979 |
| ABA 1h | 163.073 | 57.7487 | 3.44194 | 1.22626 |
| ACC 1h | 49.0708 | 13.2314 | 0.911841 | 0.241844 |
| BABA 1h | 41.9166 | 13.4594 | 0.818593 | 0.315787 |
| Chitin 1h | 24.7209 | 8.68467 | 0.512156 | 0.142368 |
| Epi 1h | 38.2551 | 8.70343 | 0.876042 | 0.218892 |
| SA 1h | 41.7328 | 6.10934 | 0.835408 | 0.11179 |
| Me-JA 1h | 21.2904 | 5.88355 | 0.509869 | 0.182246 |
| Control 6h | 35.1392 | 8.67769 | 0.680075 | 0.144297 |
| ABA 6h | 1112.05 | 92.923 | 21.8155 | 1.26128 |
| ACC 6h | 80.2131 | 6.07988 | 1.46129 | 0.109206 |
| BABA 6h | 67.6451 | 11.1021 | 1.23323 | 0.210941 |
| Chitin 6h | 47.3638 | 8.45634 | 0.900496 | 0.20978 |
| Epi 6h | 79.3471 | 11.2123 | 1.44561 | 0.110305 |
| SA 6h | 49.8616 | 19.732 | 0.812769 | 0.444475 |
| Me-JA 6h | 27.1549 | 5.18596 | 0.553696 | 0.151442 |
Source Transcript PGSC0003DMT400001688 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G80160.1 | +3 | 1e-86 | 261 | 122/156 (78%) | Lactoylglutathione lyase / glyoxalase I family protein | chr1:30151101-30151930 FORWARD LENGTH=167 |
