Probe CUST_45198_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45198_PI426222305 | JHI_St_60k_v1 | DMT400010864 | GGCTCAGATTGTCAACTGGACCTTTTACTCACAATTATGGTCTCAAATTTAGTTAATTTG |
All Microarray Probes Designed to Gene DMG400004252
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45198_PI426222305 | JHI_St_60k_v1 | DMT400010864 | GGCTCAGATTGTCAACTGGACCTTTTACTCACAATTATGGTCTCAAATTTAGTTAATTTG |
| CUST_45225_PI426222305 | JHI_St_60k_v1 | DMT400010863 | CTTGTTGTCTCTCAAACCATCAATAACTCTCAGAACGTTGGAGCTTAAAAGAAATATAGT |
Microarray Signals from CUST_45198_PI426222305
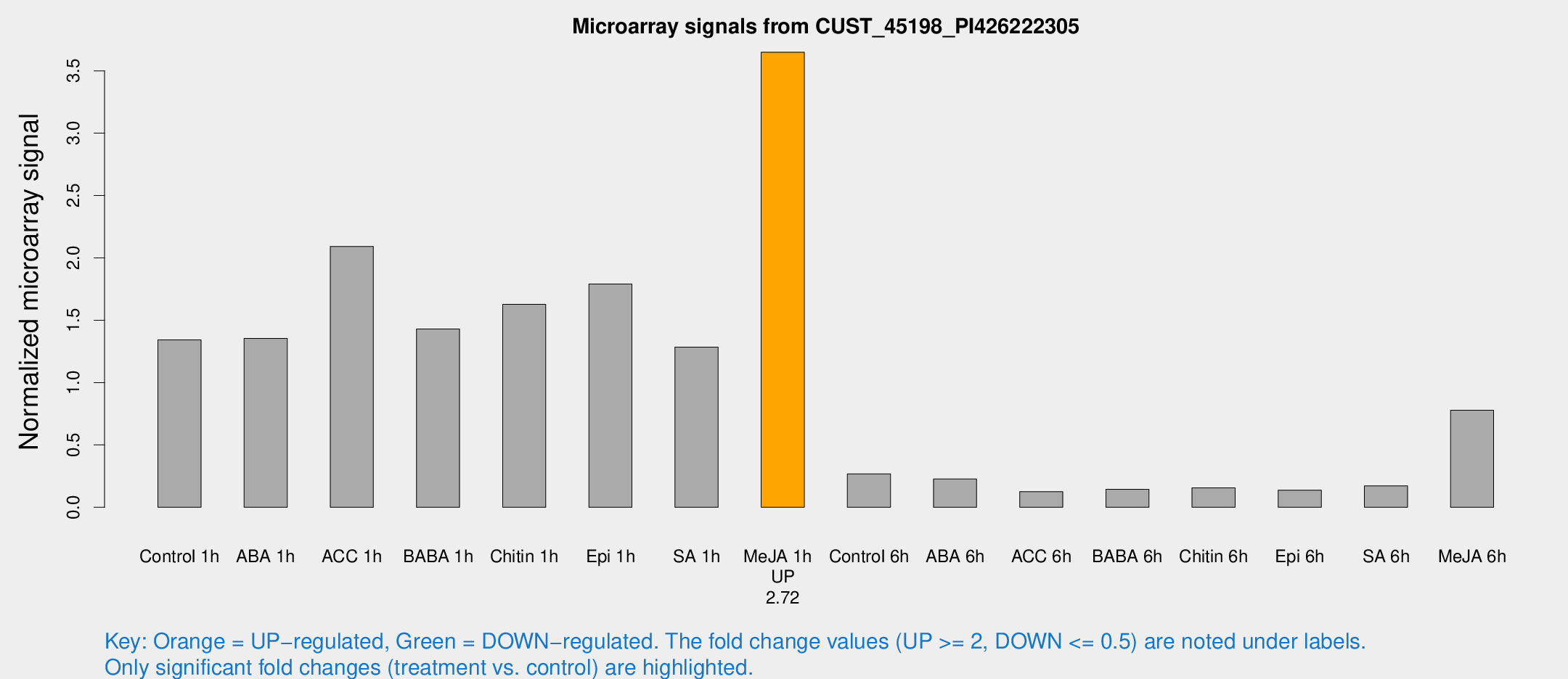
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 59.3789 | 4.62719 | 1.34334 | 0.104979 |
| ABA 1h | 55.4137 | 11.2234 | 1.35498 | 0.24292 |
| ACC 1h | 98.8505 | 19.6941 | 2.09123 | 0.315765 |
| BABA 1h | 66.4757 | 19.7885 | 1.4297 | 0.326969 |
| Chitin 1h | 66.5313 | 12.1033 | 1.62776 | 0.385257 |
| Epi 1h | 69.3322 | 9.19267 | 1.79111 | 0.259299 |
| SA 1h | 58.9401 | 7.24071 | 1.28442 | 0.132549 |
| Me-JA 1h | 131.222 | 8.21101 | 3.64929 | 0.344732 |
| Control 6h | 13.4716 | 4.2631 | 0.267797 | 0.0923644 |
| ABA 6h | 11.5205 | 3.35769 | 0.226689 | 0.0796773 |
| ACC 6h | 6.49186 | 3.86231 | 0.125438 | 0.0726799 |
| BABA 6h | 7.31451 | 3.53624 | 0.144834 | 0.0736417 |
| Chitin 6h | 7.65072 | 3.49646 | 0.156275 | 0.0777627 |
| Epi 6h | 6.84009 | 3.65342 | 0.137331 | 0.0731909 |
| SA 6h | 7.60968 | 3.33377 | 0.171449 | 0.0785122 |
| Me-JA 6h | 39.1039 | 14.3242 | 0.778468 | 0.238689 |
Source Transcript PGSC0003DMT400010864 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G22920.1 | +1 | 8e-65 | 208 | 103/185 (56%) | non-yellowing 1 | chr4:12016776-12017969 REVERSE LENGTH=268 |
