Probe CUST_43627_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43627_PI426222305 | JHI_St_60k_v1 | DMT400030786 | GTGCTAGATTGGACCATCGGAAATCTTACTTGTGTTGAAGCAAAGATTATGCTTGTCTAG |
All Microarray Probes Designed to Gene DMG400011793
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43627_PI426222305 | JHI_St_60k_v1 | DMT400030786 | GTGCTAGATTGGACCATCGGAAATCTTACTTGTGTTGAAGCAAAGATTATGCTTGTCTAG |
| CUST_43657_PI426222305 | JHI_St_60k_v1 | DMT400030787 | CACACGGACATGAAGATCAGAATGAAGTTTAAGTTTGGATTAATCTTCATTAGTACCCAA |
Microarray Signals from CUST_43627_PI426222305
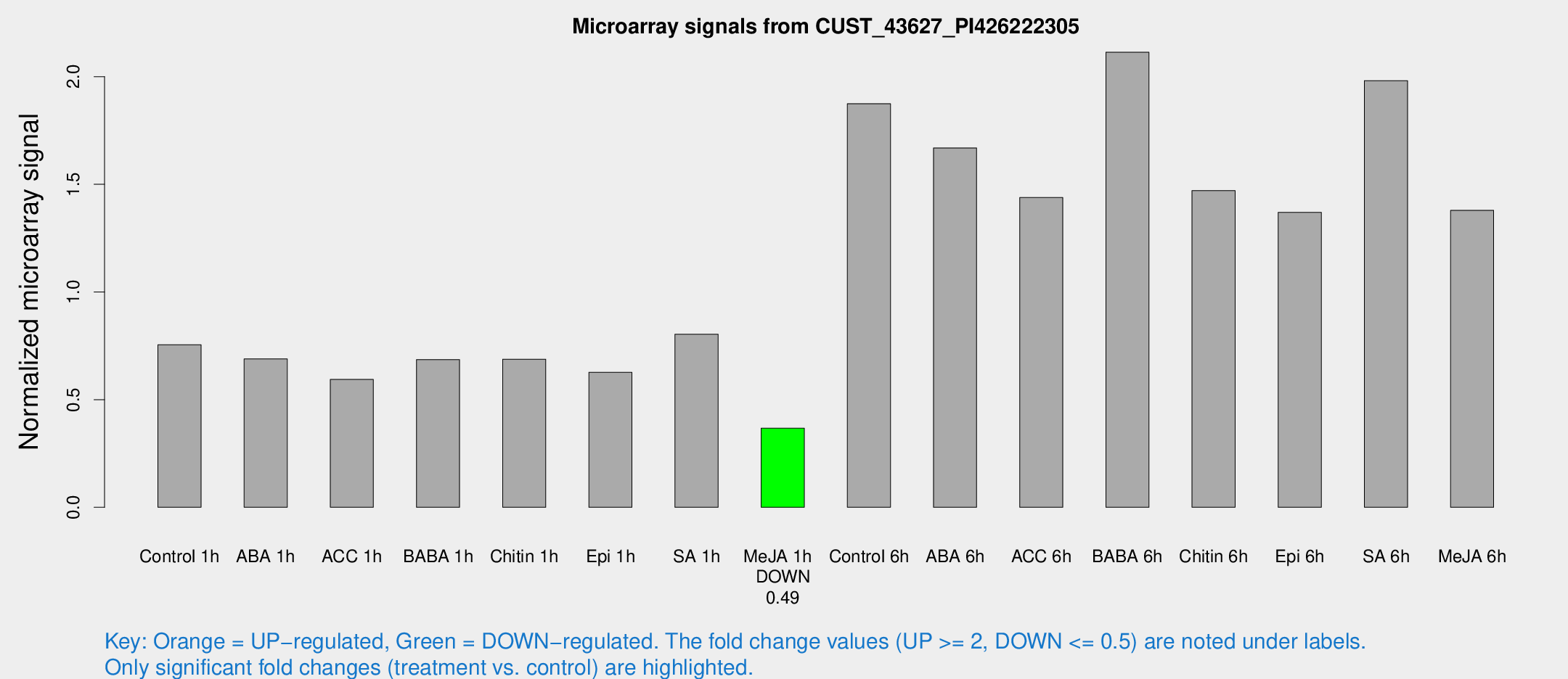
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 86.9041 | 14.3541 | 0.755004 | 0.0784771 |
| ABA 1h | 69.4559 | 8.63387 | 0.689366 | 0.107577 |
| ACC 1h | 69.2716 | 7.63864 | 0.593672 | 0.0491368 |
| BABA 1h | 77.1939 | 14.106 | 0.686269 | 0.0690044 |
| Chitin 1h | 71.2123 | 10.9247 | 0.687744 | 0.0607167 |
| Epi 1h | 63.3469 | 11.6127 | 0.627116 | 0.1195 |
| SA 1h | 94.7382 | 14.6134 | 0.803729 | 0.0804879 |
| Me-JA 1h | 33.9143 | 3.76526 | 0.367081 | 0.0411337 |
| Control 6h | 221.795 | 45.2283 | 1.87438 | 0.271835 |
| ABA 6h | 214.269 | 59.3876 | 1.6692 | 0.364439 |
| ACC 6h | 193.536 | 39.6537 | 1.43886 | 0.342176 |
| BABA 6h | 269.292 | 32.8494 | 2.11351 | 0.184213 |
| Chitin 6h | 176.317 | 10.8046 | 1.47099 | 0.0899757 |
| Epi 6h | 174.556 | 14.6187 | 1.36963 | 0.101184 |
| SA 6h | 221.623 | 19.3752 | 1.98163 | 0.190392 |
| Me-JA 6h | 165.546 | 44.0376 | 1.37925 | 0.259641 |
Source Transcript PGSC0003DMT400030786 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G21270.1 | +1 | 6e-32 | 125 | 78/222 (35%) | wall-associated kinase 2 | chr1:7444997-7447345 FORWARD LENGTH=732 |
