Probe CUST_43564_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43564_PI426222305 | JHI_St_60k_v1 | DMT400064751 | GTTTATATTGGAGAAGCTTTTGAGCTTCCATGTAATGTAGCACTTGAACCAAGTAATAAC |
All Microarray Probes Designed to Gene DMG402025150
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43565_PI426222305 | JHI_St_60k_v1 | DMT400064750 | GTTTATATTGGAGAAGCTTTTGAGCTTCCATGTAATGTAGCACTTGAACCAAGTAATAAC |
| CUST_43564_PI426222305 | JHI_St_60k_v1 | DMT400064751 | GTTTATATTGGAGAAGCTTTTGAGCTTCCATGTAATGTAGCACTTGAACCAAGTAATAAC |
Microarray Signals from CUST_43564_PI426222305
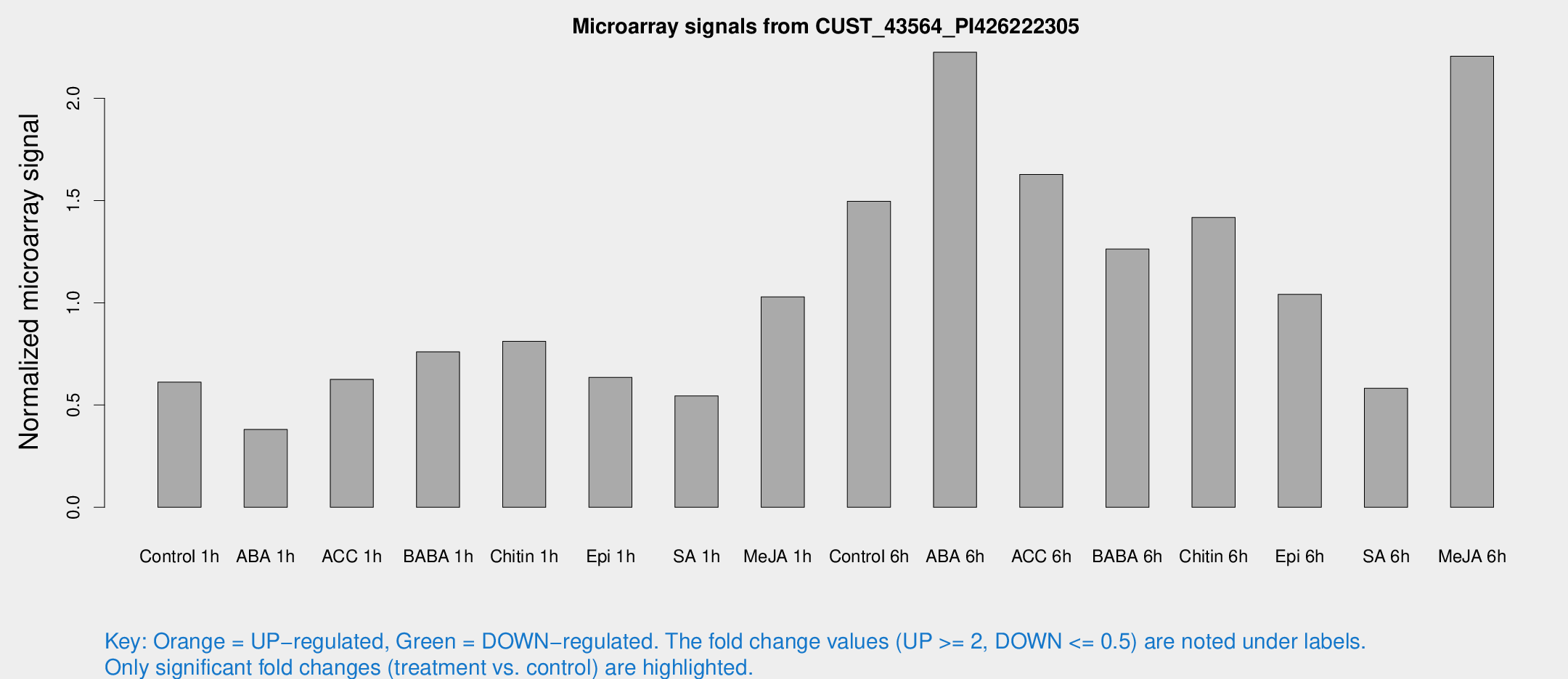
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 17.0425 | 7.48979 | 0.611927 | 0.384357 |
| ABA 1h | 8.25017 | 3.55551 | 0.380479 | 0.186214 |
| ACC 1h | 15.036 | 4.83351 | 0.625836 | 0.19429 |
| BABA 1h | 16.9779 | 4.03665 | 0.75999 | 0.185363 |
| Chitin 1h | 16.9589 | 3.78831 | 0.811561 | 0.190671 |
| Epi 1h | 13.9174 | 3.78843 | 0.635507 | 0.224923 |
| SA 1h | 18.7043 | 11.6426 | 0.544513 | 0.432763 |
| Me-JA 1h | 19.4615 | 3.975 | 1.02933 | 0.209986 |
| Control 6h | 39.8696 | 14.9635 | 1.4962 | 0.493647 |
| ABA 6h | 54.2057 | 5.0639 | 2.22516 | 0.213599 |
| ACC 6h | 42.7286 | 5.07083 | 1.62768 | 0.199519 |
| BABA 6h | 32.838 | 4.70096 | 1.26281 | 0.185429 |
| Chitin 6h | 34.7745 | 4.73128 | 1.41724 | 0.198323 |
| Epi 6h | 30.4937 | 9.32444 | 1.04129 | 0.555157 |
| SA 6h | 13.9411 | 4.19233 | 0.582209 | 0.200285 |
| Me-JA 6h | 50.481 | 4.75108 | 2.20606 | 0.209058 |
Source Transcript PGSC0003DMT400064751 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G29180.1 | +1 | 5e-167 | 499 | 265/442 (60%) | Protein of unknown function (DUF1336) | chr3:11149073-11151322 FORWARD LENGTH=513 |
